Abstract
cAMP-dependent protein kinases are reversibly complexed with any of the four isoforms of regulatory (R) subunits, which contain either a substrate or a pseudosubstrate autoinhibitory domain. The human protein kinase X (PrKX) is an exemption as it is inhibited only by pseudosubstrate inhibitors, i.e. RIα or RIβ but not by substrate inhibitors RIIα or RIIβ. Detailed examination of the capacity of five PrKX-like kinases ranging from human to protozoa (Trypanosoma brucei) to form holoenzymes with human R subunits in living cells shows that this preference for pseudosubstrate inhibitors is evolutionarily conserved. To elucidate the molecular basis of this inhibitory pattern, we applied bioluminescence resonance energy transfer and surface plasmon resonance in combination with site-directed mutagenesis. We observed that the conserved αH-αI loop residue Arg-283 in PrKX is crucial for its RI over RII preference, as a R283L mutant was able to form a holoenzyme complex with wild type RII subunits. Changing the corresponding αH-αI loop residue in PKA Cα (L277R), significantly destabilized holoenzyme complexes in vitro, as cAMP-mediated holoenzyme activation was facilitated by a factor of 2–4, and lead to a decreased affinity of the mutant C subunit for R subunits, significantly affecting RII containing holoenzymes.
Keywords: Enzyme Inactivation, Protein Kinase A (PKA), Protein-Protein Interactions, Site-directed Mutagenesis, Surface Plasmon Resonance (SPR), Bioluminescence Resonance Energy Transfer, PrKX, PrKY, αH-αI Loop
Introduction
The protein kinase A (PKA) holoenzyme is a heterotetramer composed of two catalytic (C)5 subunits kept inactive by a dimer of R subunits. Each R subunit monomer contains two tandem cAMP-binding domains, in which the sequential binding of two cAMP molecules releases an active C subunit (1). Two main classes of PKA isozymes, type I and type II, distinguishable by their R subunits, have been described. Crystal structures and solution scattering data provide evidence of a complex interaction network between C and R subunits as well as differences in global structure of PKA type I and type II holoenzymes (2–5).
Homo sapiens express the PKA R subunit isoforms RIα, RIβ, RIIα, and RIIβ and C subunits Cα, Cβ, and Cγ. Another cAMP-dependent protein kinase is PrKX and possibly protein kinase Y (PrKY). PrKY is 94% homologous to PrKX, but shortened by 81 amino acids at the C terminus. On the genome level, PRKX and PRKY are implicated in sex-reversal disorders (6). PrKX is being discussed as a phylogenetically and functionally separate enzyme (7, 8). In contrast to the ubiquitously expressed Cα subunit, PrKX is mainly active during embryonic organ development and cellular differentiation in hematopoietic lineages. It was found to be crucial for macrophage and granulocyte maturation (9, 10). PrKX was shown to be involved in renal development, regulating epithelial cell migration, ureteric bud branching, and induction of glomeruli formation (8, 11–13).
Cα and human PrKX differ by their selective holoenzyme formation in living cells, as PrKX is inhibited only by RIα, but not by RIIα (14, 15). Here, we have tested all four human R subunits for the first time side by side and show that this so far unique property of RI over RII preference with respect to autoinhibition appears to be an evolutionarily conserved feature of PrKX and at least four of its orthologs (Mus musculus Pkare, Drosophila melanogaster DC2, Trypanosoma brucei PKAC3, human PrKY), and possibly also Caenorhabditis elegans F47F2.1b.
Previously, we identified the R subunit autoinhibitory site as a main determinant for isoform-specific regulation of PKA (15). We were able to gain significant binding of PrKX with RIIα solely by mutation of Ser-99 to Ala in the RIIα autoinhibitory domain (P0-site). Conversely, introducing an autophosphorylation site Ser or an Asp in the inhibitory domain of RIα completely abolished binding to PrKX, but not to Cα. This led us to believe that at least one peripheral interaction interface could be non-functional in PrKX holoenzymes when compared with the Cα holoenzymes. We therefore set out to pinpoint this position in PrKX, applying mainly BRET-based cell interaction assays and SPR analyses with wild type and mutant C subunits. Our investigations led to the identification of residue Arg-283, located in the PrKX αH-αI loop, as important for the differential regulation of the PrKX subfamily, i.e. its selective autoinhibition by RI subunits in living cells. Supported by biochemical and modeling evidence we provide evidence that an Arg at position 283 in PrKX or at position 277 in the mutant Cα interferes with holoenzyme stability and allosteric regulation of the kinases by disturbing a crucial interaction platform composed of activation loop (C subunit) and αA-helix (R subunit) residues.
EXPERIMENTAL PROCEDURES
Protein Expression Vectors
The oligonucleotides and vectors used for cloning of prokaryotic and eukaryotic expression vectors and for subsequent site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) are provided under supplemental Table S2. Cloning of human PRKAR1A, PRKAR2A, PRKACA, and PRKX genes into the BRET vectors was published previously (15, 16). Mutagenesis of PrKX was performed using plasmid pFastBac HTb-PRKX (15) as the template. For protein purification, human Cα was subcloned into pHis5BA via pRSETB-hCα (supplemental Table S2). Pkare was amplified from pCMVSport6-Pkare (RZPD, Berlin, Germany). PRKY was amplified from cDNA clone pCMV6-XL5-PRKY (accession number NM_002760.2) purchased from OriGene (Rockville, MD). C. elegans F47F2.1b was amplified from pDONR201-F47F2.1b, provided by Dr. M. Jedrusik-Bode, MPI for Biophysical Chemistry, Göttingen, Germany. A DC2 cDNA clone (FlyBase clone number AT10577) was obtained from the Drosophila Genomics Resource Center (Bloomington, IN). The full-length clone and a 1–227 deletion variant were amplified and each subcloned into GFP2-C2. The T. brucei PKAC3 coding region (UniProt accession number Q388U5)6 was amplified to allow subcloning into GFP2-C3.
Plasmids for expression of R subunits RIα (pRSETB-PRKA1A) and RIIα (pRSETB-PRKA2A) were a kind gift from Prof. Dr. S. S. Taylor, University of California, San Diego. pRSETB-PRKA1B and Rluc(h)-N2-PRKA1B were subcloned from pBluescript-PRKA1B, a gift of Prof. Dr. K. Tasken, University of Oslo, Norway. The human PRKA2B coding region was amplified from a cDNA clone (OriGene, accession number NM_002736.2) and subcloned in Rluc(h)-N2 and pRSETB, respectively. Point mutations of Cα, PrKX, F47F2.1b, PKAC3, RIβ, and RIIβ were introduced using the oligonucleotides listed under supplemental Table S2. Mutagenesis of RIα and RIIα was published previously (15). All plasmids used in this study were sequence verified.
BRET Assays
COS-7 cells (ATCC CRL-1651) were transfected in white 96-well microplates (Nunc) as described before (17) using 0.25 μg of DNA per plasmid and well. Two days after transfection, cells were rinsed with glucose-supplemented Dulbecco's PBS (D-PBS, Invitrogen), and treated with forskolin (50 μm) and 3-isobutyl-1-methylxanthine (IBMX) (500 μm) at final concentrations in D-PBS for 20 min. Coelenterazine 400a (Biotrend) was added immediately before BRET read-out. The light output was taken consecutively (read time 1 s, gain 25) for each well with filters at 410 nm wavelength (±80 nm band pass) for the Renilla luciferase (Rluc) and 515 nm (±30 nm band pass) for the GFP2 emission using an α-fusion microplate reader (PerkinElmer Life Sciences). Wells containing cells expressing Rluc alone were included in every experiment (background BRET signal, bg). Emission values (em) obtained with untransfected (n.t.) cells were subtracted, and BRET ratios were calculated as follows: (em515 nm − n.t. cells515 nm)/(em410 nm − n.t. cells410 nm). In general, experiments were repeated at least three times with six wells per experimental condition. Average results are represented as mean ± S.E. Statistical evaluation (one-way analysis of variance with Newman-Keuls post tests) was carried out with the GraphPad Prism software version 4.0 (GraphPad).
Protein Expression and Purification
Expression and affinity purification of R subunits was carried out as described (18, 19). His6-tagged human Cα (His6-Cα) and His6-CαL277R were expressed overnight at room temperature in Escherichia coli BL21(DE3) (Novagen) and purified using a Talon affinity resin and standard conditions (Clontech). Expression and purification of His6-PrKX and His6-PrKXR283L was performed as described (15).
Spectrophotometric Kinase Activity Assay
The specific activity of the recombinant PKA Cα was tested by the continuous enzyme-linked spectrophotometric method described by Cook et al. (20) using 260 μm of the synthetic substrate Kemptide (LRRASLG; Biosynthan). 1 Unit/mg is defined as 1 μmol × min−1 × mg−1. Apparent activation constants (Kact) were determined with 10 or 20 nm reconstituted holoenzyme by adding varying concentrations of cAMP in assay buffer containing 1 mm ATP and 10 mm MgCl2 (15).
Surface Plasmon Resonance
Methods for interaction analyses of Cα and PrKX with R subunits were published previously (15). Briefly, 200–300 resonance units (RU) of His6-Cα, His6-CαL277R, His6-PrKX, and His6-PrKXR283L were covalently coupled on a modified Ni2+-nitrilotriacetic acid (NTA) chip via primary amines. A dilution series (0.5 to 256 nm R subunit) was injected at a flow rate of 30 μl/min, and association and dissociation were recorded for 5 min each. A blank NTA surface was used as the control surface. Additionally, an injection of buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 0.005% Tween 20, 50 μm EDTA, 1 mm ATP, and 10 mm MgCl2) was subtracted from each sensorgram. Surface regeneration was achieved by injecting 100 μm cAMP and 2.5 mm EDTA, diluted in running buffer. The rate constants for association (ka) and dissociation (kd) were fitted assuming a 1:1 Langmuir binding model using Biaevaluation 4.1 (Biacore AB). Equilibrium binding constants (KD) were calculated by dividing kd with ka.
Molecular Dynamics (MD) Simulations
The structural models of Cα and CαL277R in complex with the bovine (b) RIIα were based on the available x-ray structures solved at 2.5 Å of resolution (Protein Data Bank code 2QVS (4)). The L277R mutant side chain was modeled to maintain the orientation of the wild type side chain in the experimental structures. Missing atoms were added assuming standard bond lengths and angles. The models were immersed in parallel piped water boxes, whose edges were ∼9, 10, and 12 nm; sodium or chlorine ions were added to neutralize the boxes. MD simulations were performed using the computational setup already applied before (21, 22), whose main properties are: the MD program Gromacs was employed (23), using the Amber99 force field for protein and TIP3P for water (24, 25), Lincs algorithm to constrain bonds (26), periodic boundary conditions, 10 Å cutoff for short range electrostatic and van der Waals interactions, particle mesh Ewald to treat long range electrostatics (27), and 2-fs integration time step. After energy minimization, 300 ps of MD of the solvent with gradual heating of the systems, the complexes underwent 14 ns of MD in the NPT ensemble at 300 K and 1 atm pressure, using the Berendsen thermostat and barostat (28). Root mean square deviations were calculated on α-carbon atoms using the initial conformation as reference. Solvent accessible surfaces are calculated for the residues of the four complexes using the center-probe area definition (29), as in Ref. 30, and averaged over the second halves of the MD trajectories.
RESULTS
Inhibition of PrKX and Orthologs by Human R Subunits
Previous in vitro and BRET-based interaction analyses in living cells using RIα and RIIα subunits identified human PrKX as a type Iα-specific PKA-like kinase (14, 15). To study binding patterns of PrKX-like kinases, we investigated binding of all four human R isoforms to PrKX, human PrKY, murine Pkare (31, 32), D. melanogaster DC2 (33), and a putative PrKX homolog from the protozoan parasite T. brucei (PKAC3, GenBank accession number AF253418 (34)).6 They were all cloned as N-terminal GFP2 fusions for eukaryotic protein expression. In analogy to the previously published protein expression constructs for human RIα and RIIα (16), Renilla luciferase (Rluc) was genetically fused to the C terminus of RIβ and RIIβ.
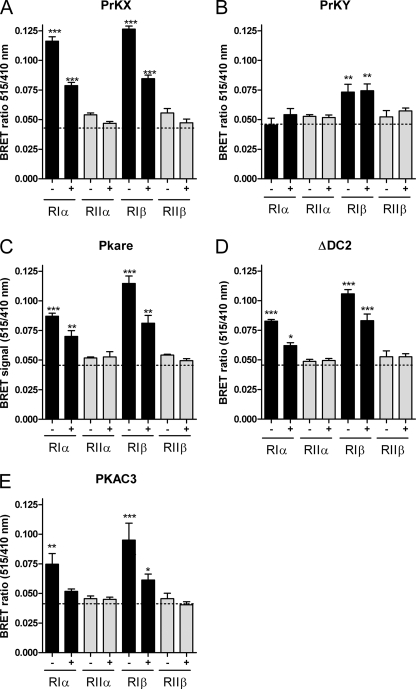
For BRET analyses in COS-7 cells, the five investigated PrKX-type kinase fusion constructs were co-transfected with each of the four human R subunits at a 1:1 ratio of plasmid DNA. Employing this BRET assay, a signal above the background indicates PKA holoenzyme formation, which can be reduced by increasing intracellular cAMP ([cAMP]i). Furthermore, the BRET signal of a given interaction pair provides a quantitative measure of the amount of holoenzyme present in resting versus stimulated cell populations (17). Fig. 1 shows the protein-protein interaction analyses, where PrKX (A), PrKY (B), Pkare (C), a deletion variant of DC2 (δ1–227, ΔDC2) (D) and PKAC3 (E) were each probed by BRET for binding to the four human R subunits. With DC2 we achieved BRET signals only with the N-terminal deletion variant, but not the wild type protein (data not shown). The BRET analyses revealed that PrKX, Pkare, ΔDC2, and PKAC3 displayed very similar R subunit binding patterns, interacting specifically only with RIα and RIβ. In all cases, we observed the highest overall BRET values when RIβ was co-expressed with the C subunits. PrKY (Fig. 1B) binds only to RIβ resulting in a BRET signal of about 30% of the PrKX:RIβ signal (compare Fig. 1, A and B). Binding of RI to the C subunits was reduced by ∼50% upon treatment with 50 μm forskolin and 500 μm IBMX, which we used to get to a sustained increase of [cAMP]i, in agreement with previous results (Fig. 1) (15). Independent investigations showed that (local) substrate availability greatly enhances the sensitivity of PKA type Iα toward [cAMP]i (35, 36). In contrast, the PrKY/RIβ interaction was not influenced by [cAMP]i (Fig. 1B), indicating that not only R subunit docking, but also its release could be hampered in PrKY. This result suggests that PrKY is lacking R interaction interface(s). In contrast to PrKX, we did not detect BRET with PrKY and any of the human isoforms of the heat stable protein kinase inhibitor (PKI (37) (data not shown). Attempts to purify active His6-PrKY after heterologous expression in Sf9 cells were also unsuccessful. We can therefore conclude that the C-terminal deletion of the kinase, which includes the last domain of the conserved catalytic core (subdomain 11) (38), likely leads to the expression of an inactive enzyme with a significantly diminished binding of substrates and pseudosubstrates.
FIGURE 1.
PKA RI-specific interaction of PrKX-like kinases is conserved between species. To follow PKA subunit interaction in living cells, PrKX (A), PrKY (B), Pkare (C), ΔDC2 (D), and PKAC3 (E) were each fused N-terminally with GFP2. RIα, RIIα, RIβ, and RIIβ were cloned into eukaryotic vectors fusing them C-terminally with Renilla luciferase (Rluc). For BRET experiments, COS-7 cells were co-transfected with the indicated constructs and grown for 2 days prior to the incubation with vehicle (−) or with (+) 50 μm forskolin, 500 μm IBMX dissolved in Dulbecco's PBS. BRET was measured immediately after addition of the luciferase substrate. Values are given as mean ± S.E. of at least three independent experiments, each performed with n = 6 replicates (***, p < 0.001; **, p < 0.01; *, p < 0.05). The dotted line indicates the mean background value.
In contrast to PrKX and PrKX-like kinases, human Cα, Cβ1, Cβ2, and Cαs interact with all four human R subunits based on BRET (15, 16).7 Thus, data provided here support an RI selective holoenzyme formation as a main characteristic of PrKX and four of its orthologs.
Influence of the αH-αI Loop of Cα and PrKX on Differential R Subunit Binding
To investigate inhibition of PrKX and conventional PKA C subunits in more detail, we aligned the sequences of the PKA-related kinase subfamilies and subjected them to phylogenetic analysis (supplemental Figs. S1, A–C, and Table S1). From x-ray structural analysis of either RIα or RIIα in complex with the Cα subunit, the αH-αI loop region of the C subunit was proposed to be important particularly for binding of the RIIα subunit (3, 4). Residues Asp-276Cα and Thr-278Cα as well as the residues Thr-278Cα and Lys-285Cα were previously found to interact with the cAMP binding domain A of bovine RIα (Arg-352, Arg-355) and bovine RIIα (Arg-365, Leu-266) subunits (Fig. 2A). These αH-αI loop residues are conserved between PKA and PrKX-like kinases. To elucidate residues necessary for the interaction with RII subunits, we first mutated Lys-285 to Ala in Cα and monitored holoenzyme formation using the BRET assay. This mutation had no effect on C subunit binding to any of the four human R subunits (data not shown), in agreement with a previous study investigating a Lys-285 to Pro mutant (39), suggesting a backbone rather than a side chain interaction between Lys-285Cα and Leu-366bRIIα.
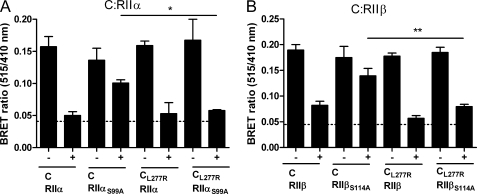
FIGURE 2.
Mutant PrKX (R283L) has the capacity for holoenzyme formation with RII subunits. A, this partial alignment of the αH-αI loop region of Cα and PrKX-like kinases highlights the conserved Cα residues previously found to be involved in bRIα (R352, R355) and bRIIα (R365, L366) subunit coordination as well as intramolecular stabilization of Cα (E208Cα), all shaded in black. Non-conserved and/or experimentally addressed residues in this study are shaded gray and written in black (L277Cα), red (R283PrKX, R280Pkare, R324F47F2.1b, R510DC2), and yellow (P254PKAC3, E253PKAC3). B, BRET interaction analyses of wild type and PrKXR283L with wild type or P0-site mutant R subunits were carried out as detailed in the legend to Fig. 1. Plasmids coding for GFP2-PrKX and GFP2-PrKXR283L were co-transfected with constructs expressing either RIα-Rluc or RIαA99S-Rluc (C), as in B, co-transfected with either RIβ-Rluc or RIβG99S-Rluc constructs. D, as in B, combined with either RIIα-Rluc or RIIαS99A-Rluc. E, as in B, combined with either RIIβ-Rluc or RIIβS114A-Rluc. Depicted are original BRET values (mean ± S.E.) obtained from at least three independent repeats (n = 6 wells; ***, p < 0.001; **, p < 0.01; *, p < 0.05). + indicates treatment with forskolin/IBMX, − indicates mock treatment, and the dotted line represents the mean background value.
We then focused on a αH-αI loop residue that differs between PrKX and Cα, Arg-283PrKX and Leu-277Cα. Arg-283PrKX is conserved in all but one of the PrKX-like kinases investigated in this study and the corresponding residue Leu-277Cα is common to Cα and -orthologs (Fig. 2A and supplemental Fig. S1, B and C). We went on to replace Arg-283PrKX by Leu in PrKX and investigated the mutant protein for specific activity, intracellular and in vitro binding to wild type and P0-site mutant R subunits. The activities of the purified PrKX proteins were tested with a coupled spectrophotometric assay using the synthetic substrate Kemptide (LRRASLG) (20). Results were almost identical for PrKX (14) and mutant PrKXR283L (1.4 ± 0.2 units/mg, data not shown).
As depicted in Fig. 2, D and E, changing Arg-283PrKX to Leu had a significant stabilizing effect on wild type RIIα and RIIβ subunit binding in living cells. SPR analyses using the corresponding purified proteins lead to an about 10–12-fold increase in binding affinity of PrKX to RIIα subunits (Table 1 and Fig. 3, C and D). A combined expression of PrKXR283L and P0-site mutants RIIαS99A or RIIβS114A, lead to BRET signals comparable with RIα and RIβ wild type holoenzymes (Fig. 2). Using SPR, a KD value of 251 nm compared with ∼3–4 μm (Table 1) translates into a ∼12–16-fold increased affinity of PrKXR283L and RIIαS99A. As depicted in Fig. 2, B and C, results of BRET assays employing either RIα or RIβ in combination with wild type PrKX or PrKXR283L display a similar holoenzyme dynamics. As previously shown for RIα wild type holoenzymes with Cα or PrKX (15, 16), wild type RIβ holoenzymes with PrKX and PrKXR283L do not respond with complete dissociation upon cAMP elevation in living cells (Fig. 2, B and C). In vitro, the affinity of RIβ and PrKXR283L measured by SPR increased by about a factor 3 (Table 1 and Fig. 3B), due to a reduction in the off-rate. The interaction of wild type PrKX with mutant RIα and RIβ resulted in comparably high BRET values (Fig. 2, B and C and Ref. 15). Introducing a Ser residue at the P0-site in both type I R subunits prohibited binding of PrKX. In the case of RIα, but not RIβ, the negative effect of this mutation was partially compensated by changing Arg-283PrKX to Leu (Fig. 2, B and C). This was also reflected by a 2-fold stronger in vitro binding of RIαA99S to PrKXR283L compared with PrKX (Table 1). In a previous study, we showed that interaction of PrKX with an in vitro phosphorylated RIαA99S protein was completely abolished (15). From this we can conclude that Arg-283PrKX located in the αH-αI loop region prohibits high-affinity binding to and thus functional inhibition of PrKX by wild type RII or mutant RI subunits in living cells.
TABLE 1.
Rate and equilibrium binding constants of PrKX/R and Cα/R subunit interaction derived from SPR analyses
Concentration series of the respective R subunit (0.5–512 nm) were injected over surfaces containing 200–300 response units covalently immobilized to PrKX, PrKXR283L, Cα, or CαL277R in the presence of ATP/Mg2+. The association (ka) and dissociation (kd) rate constants were determined by global fitting assuming a 1:1 Langmuir binding model utilizing Biaevaluation 4.1 (Biacore AB). Equilibrium binding constants (KD) were calculated by dividing kd with ka.
| RIα | RIβ | RIIα | RIIβ | RIαA99S | RIIαS99A | |
|---|---|---|---|---|---|---|
| PrKX | ||||||
| ka (m−1 s−1) | 2.6 × 105 | 6.9 × 105 | 2.3 × 105 | 2.1 × 104 | ||
| kd (s−1) | 2.2 × 10−4 | 2.8 × 10−3 | 1.1 × 10−2 | 4 × 10−3 | ||
| KD (nm) | 0.8 | 4 | ∼3–4 μm | ∼5–10 μm | 47 | 195 |
| PrKXR283L | ||||||
| ka (m−1 s−1) | 5.9 × 105 | 7.9 × 105 | 2 × 105 | 3.2 × 104 | 7.3 × 105 | 5.1 × 104 |
| kd (s−1) | 5 × 10−4 | 1.1 × 10−3 | 5.7 × 10−2 | 2.0 × 10−2 | 1.4 × 10−2 | 1.3 × 10−2 |
| KD (nm) | 0.8 | 1.4 | 285 | 650 | 19 | 251 |
| Cα | ||||||
| ka (m−1 s−1) | 8.1 × 105 | 2.2 × 106 | 3.1 × 105 | 3.9 × 105 | 8.6 × 105 | 2 × 105 |
| kd (s−1) | 8 × 10−5 | 3.8 × 10−4 | 2.3 × 10−4 | 3.1 × 10−4 | 1 × 10−3 | 4.6 × 10−4 |
| KD (nm) | 0.1 | 0.17 | 0.77 | 0.79 | 1.2 | 2.3 |
| CαL277R | ||||||
| ka (m−1 s−1) | 9.1 × 105 | 2.4 × 106 | 5 × 105 | 3 × 105 | 9.7 × 105 | 1.6 × 105 |
| kd (s−1) | 1.9 × 10−4 | 7.8 × 10−4 | 2.6 × 10−3 | 1.1 × 10−3 | 1.4 × 10−3 | 1 × 10−3 |
| KD (nm) | 0.2 | 0.33 | 5.2 | 3.5 | 1.4 | 6.2 |
FIGURE 3.
Representative SPR sensorgrams of R subunit binding to immobilized His6-tagged PrKX, PrKXR283L, Cα, and CαL277R. All C subunits were covalently coupled on Ni2+-NTA sensor surfaces. Shown are normalized binding curves, where 128 nm of each RIα (A and E), RIβ (B and F), RIIα (C and G), and RIIβ subunit (D and H) were injected at a flow rate of 30 μl/min in running buffer containing ATP/Mg2+, except for PrKX, where an injection of 512 nm of the RIIα/RIIβ subunits is depicted (C and D). Insets in panels C and D represent original SPR data for the PrKX/RIIα and PrKX/RIIβ interactions.
Initial investigation of a putative ortholog of PrKX from C. elegans (F47F2.1b) did not lead to significant interaction with human R subunits in the BRET assay. We therefore reasoned that replacing the Arg at position 324 for Leu in F47F2.1b might stabilize the interaction with R subunits in analogy to human PrKX. This was indeed the case, as we gained interaction of the mutant C subunit with RIα and RIβ subunits (supplemental Fig. S2A). The PKAC3 protein from T. brucei is the only putative PrKX ortholog investigated in this study that does not carry an Arg at the position corresponding to Arg-283PrKX but a Pro (Fig. 2A). We therefore investigated the interaction of the protein to R subunits after replacing Pro-254PKAC3 with Leu. Interestingly, interactions of wild type PKAC3 with either RIα or RIβ subunits were reduced to background levels upon mutation of the C subunit, indicating the importance of the Pro residue for holoenzyme formation with this T. brucei C subunit (supplemental Fig. S2B).
Mutation of Leu-277Cα Influences Holoenzyme Activation
To test the hypothesis of a central role of Leu277Cα, which corresponds to Arg-283PrKX, in maintaining a stable PKA holoenzyme complex, we investigated the consequence of introducing an Arg at position 277 in Cα in vitro and in living cells. This mutation had no effect on the specific activity of the purified kinase, determined by the coupled spectrophotometric assay (20) (20 ± 1.1 units/mg, data not shown). We then tested the activation of reconstituted holoenzymes containing wild type and CαL277R with four R isoforms in vitro (Table 2). Intriguingly, the apparent activation constants (Kact) of all mutant holoenzymes tested were significantly reduced, in other words, the activation threshold for cAMP is lowered in the mutant holoenzyme complexes under in vitro conditions. The effects ranged from a reduction by factor 2 in the case of RI subunits up to factor 4 in the RIIβ containing holoenzyme. These results were obtained with proteins from two independent protein preparations.
TABLE 2.
Apparent activation constants (Kact) are reduced in mutant (CαL277R) holoenzymes
Activation constants for cAMP were determined using a spectrophotometric kinase activity assay (20), employing the substrate peptide Kemptide and 20 nm (10 nm for RIβ) mutant or wild type holoenzyme per experimental condition. Holoenzymes were formed in a buffer containing Mg2+ ATP by adding 1.2-fold molar access of purified R subunits to the C subunits, as indicated, and incubated with a dilution series of cAMP, to obtain Kact values. The data are derived from four experiments, each performed in duplicate. They are given as mean ± S.D.
|
Kact for cAMP (±S.D.) |
||||
|---|---|---|---|---|
| RIα | RIβ | RIIα | RIIβ | |
| nm | ||||
| Cα | 105.5 ± 7 | 33.5 ± 0.7 | 118 ± 3 | 405 ± 14 |
| CαL277R | 55 ± 4 | 16 ± 7 | 53 ± 5 | 108 ± 16 |
Interaction analyses by SPR revealed that the affinity of the CαL277R subunit toward R subunits is reduced (Table 1 and Fig. 3, E–H). The effect is again more evident with RII subunits (factor 6.75 for RIIα, factor 4.4 for RIIβ), compared with a factor 2 for RIα and RIβ. The increased KD values were due to faster off-rates with the CαL277R compared with wild type Cα (Table 1 and Fig. 3, E–H). We then went on to test the interaction of PKA Cα as well as CαL277R with wild type and mutant human R subunits in living cells using the BRET assay. In accordance to a previous study comparing the PKA-type Iα and PKA-type IIα holoenzyme dissociations upon cAMP elevation, we observed that RIα or RIβ containing holoenzymes are less sensitive to activation by cAMP (Ref. 15, and data not shown), in contrast to the RIIα and RIIβ containing holoenzymes (Fig. 4, A and B), which readily activate. This almost dominant inhibitory effect of the RI subunits is lost by simple mutation of the autoinhibitory site from a pseudosubstrate to a substrate site (Ref. 15, and data not shown). In contrast, replacing the RIIα or RIIβ subunit P0-site Ser with Ala prohibits a complete activation by cAMP in the context of the living cell (Fig. 4, A and B). Interestingly, this inhibitory effect of the P0-site mutation was lost, and [cAMP]i was again able to activate the holoenzymes significantly when the RIIα/β subunits were combined with the mutant CαL277R (Fig. 5). In the case of the PKA type I holoenzymes, mutation of the C subunit αH-αI loop had no measurable effect on holoenzyme formation or dissociation in living cells (data not shown).
FIGURE 4.
Mutation of Cα subunit influences intracellular holoenzyme dynamics. BRET analyses were carried out as detailed in the legend to Fig. 1. A, plasmids coding for GFP2-Cα and GFP2-CαL283R were co-transfected with constructs expressing either RIIα-Rluc or RIIαS99A-Rluc. B, as in A, combined with either RIIβ-Rluc or RIIβS114A-Rluc constructs. Depicted are original BRET values (mean ± S.E.) obtained from at least three independent repeats (n = 6 wells; **, p < 0.01; *, p < 0.05). +, indicates treatment with forskolin/IBMX; −, indicates mock treatment. The dotted line represents the mean background value.
FIGURE 5.
Illustration of the intersubunit interface around Leu-277Cα of the PKA type IIα holoenzyme. Cα and bRIIα subunits are drawn in traces, green and red, respectively. The snapshots were taken at the end of MD simulations. A, wild type PKA Cα; and B, PKA mutant CαL277R in complex with bRIIα. Residues involved in intersubunit interaction site 3 (4) and discussed in the text are drawn in sticks and are colored by atom type; in particular, Leu277Cα (Arg) is drawn in ball and sticks format. For clarity, hydrogen atoms are not shown. Dashed lines indicate salt bridge interactions. Molecular surfaces for Lys-192 and Arg-194 of Cα, for Ser-264 and Met-267 of bRIIα are drawn in gray.
In summary, the mutation of Leu-277 in Cα leads to destabilization of the PKA holoenzyme in vitro, due to a reduced subunit affinity and activation at lower cAMP concentrations. This effect is stronger when investigating holoenzyme formation with RII subunits, thus resembling the situation in PrKX holoenzymes, which exhibit a strongly reduced binding affinity in vitro leading to no significant RII subunit binding in vivo. Under steady-state conditions in living cells, mutation of the C subunit at position Leu-277 to Arg has more subtle effects, influencing the stability of RII containing holoenzymes.
Modeling of the Cα Leu-277 to Arg Mutation
To gain insight in putative structural and regulatory consequences of changing Cα Leu-277 to Arg on the interaction network of PKA type II, we modeled the wild type and mutant complexes on the basis of the available experimental structure of the PKA RIIα·Cα complex (PDB entry code 2QVS (4)). The model was refined by means of MD simulations: 14 ns of MD were performed and root mean square deviations fluctuated at about 3 Å. In the wild type structure, Leu-277Cα is located in the intersubunit cleft, packed against the side chain of Arg-280Cα, which forms a salt bridge interaction with Glu-208Cα (40). On the other side of Leu-277Cα, the so called intersubunit interface site 3 (3, 4) includes a salt bridge network of the activation loop and αA-helix residues formed by Arg-194Cα, Asp-271bRIIα, and Arg-245bRIIα (Fig. 5A). This region is crucial for stabilizing the holoenzyme conformation as well as for the allosteric coupling in the PKA holoenzyme. For instance, the Arg-245/Asp-271bRIIα salt bridge plays a key role in coupling the cAMP binding domains of the R subunit (3, 41). In the complex, the tip of the activation loop of Cα, namely residues Lys-192-Gly-193-Arg-194, is packed in the intersubunit interface to form hydrophobic contacts with Ser-264 and Met-267 in bRIIα (Fig. 5A). These interactions are observed in the experimental structures and are reproduced in our simulations of the wild type complex. On the contrary, mutating Leu-277Cα to Arg affects the conformation of the Asp-271bRIIα side chains, which rotate toward and interact with Arg-277Cα (Fig. 5B). The interaction network of the site is affected so that the solvent exposition of the residues in the tip of the activation loop increases by about 45 Å2 in the mutant complex, with respect to the wild type, indicating higher solvation of the intersubunit interface. Although the KD values of the wild type versus mutant type IIα holoenzyme decreased only by a factor of 6.75, still being in the nanomolar range of affinity (Table 1), the theoretical structural analysis supports our experimental data, which showed a decrease in binding affinity of RII subunits to CαL277R and a facilitated activation of the mutant PKA holoenzymes.
DISCUSSION
Phylogenetic analyses performed on the kinase core (8) and the overall sequence of PrKX (supplemental Fig. S1, A and B, and Table S1) indicate that the kinase and its orthologs belong to the family of AGC kinases but are distinct from conventional PKA. Although the kinase core of PrKX shares significant overall homology to PKA Cα (∼57% identity (8)), other regions, including the N and C termini (∼23 and ∼40% identity, respectively) have diverged. The N terminus is the least conserved part, even among PrKX-like kinases. Large insertions, for example, in DdPK2 from Dictyostelium discoideum (42, 43), and in DC2 from D. melanogaster (33) are characteristic for some PrKX-like kinases (supplemental Fig. S1A). This diversity is reflected in the phylogenetic analysis of the overall sequence (supplemental Fig. S1A), the kinase core, and the C termini of the kinases (not shown), where PKA C subunits are placed in a well resolved group, clearly separated from other kinase families. On the contrary, a PrKX subfamily cannot be as clearly drawn by phylogenetic analysis only. Taking into account the experimental data provided herein, we would like to propose a more conserved PrKX subfamily including vertebrate and insect PrKX and a more extended family, which includes C. elegans (F47F2.1b), T. brucei (PKAC3), and possibly D. discoideum (DdPK2) (supplemental Fig. S1A). Unfortunately, all attempts to express full-length DdPK2 or an N-terminal deletion variant (Δ1–136) for interaction analyses in mammalian cells were unsuccessful (data not shown).
To address the PrKX family experimentally, we focused on its conserved R subunit inhibitory pattern, and we are able to provide evidence that human PrKX, mouse Pkare, D. melanogaster ΔDC2, and T. brucei PKAC3 all share the common feature of a RI over RII subunit preference with respect to autoregulation (Fig. 1). Human PrKY, which lacks the αH-αI loop, and a mutant C. elegans kinase (F47F2.1bR324L), bind only to RIβ when tested with BRET (Fig. 1 and supplemental Fig. S2). By this, we conclude that many if not all kinases of the PrKX family are autoinhibited only by RI subunits, which contain a pseudosubstrate autoinhibitory domain. To substantiate these results, interaction analyses with homologous R subunits have to be performed. Recently, the D. melanogaster protein Swiss cheese was identified as a non-canonical R subunit binding to DC2, inhibiting both DC2 and the esterase activity of Swiss cheese mainly in the brain (44). Whether this regulation is conserved in mammalian tissues, awaits experimental evaluation, but this also points to the functional separation of conventional PKA and PrKX-like kinases.
Comprehensive mutational analyses on yeast PKA by Gibbs et al. (41) and x-ray structural analyses (3, 4) suggest that in addition to the active site of the kinase at least two more interaction sites with R subunits are present that, in combination, appear to mediate high-affinity binding of PKA C and R subunits. However, we and others (15, 35, 45) demonstrated that RII subunits, due to autophosphorylation of their inhibitory domain, do not bind with high affinity to the active site cleft of C subunits. It was proposed that binding of the RIIα subunit αB-helix to the αH-αI loop of the kinase is necessary for high affinity interaction of this R subunit to conventional C subunits (4, 46) (Fig. 2). We now show that the inability of RII subunits to regulate PrKX is conferred by a combination of both, the type II R subunit autoinhibitory domain and the PrKX Arg-283 residue located on the αH-αI loop (Fig. 2). Either mutating the autoinhibitory site in RII to a pseudosubstrate, or changing the Arg-283PrKX to the invariant Leu present in conventional C subunits, reconstitutes significant binding of RII subunits to PrKX (Fig. 2). A combination of both mutations did not result in additive, subnanomolar affinities in the SPR assay (Table 1). However, the affinities of RIα/RIβ interactions with wild type PrKX are already 8- and 24-fold lower compared with the corresponding Cα interactions (Table 1). It is conceivable that other non-conserved amino acid substitutions in the kinase core, especially Asp-199PrKX (Gly-193Cα) or Asn-140PrKX (Arg-134Cα) might account for the overall lower R subunit binding affinities of PrKX and PrKXR283L (8, 41).
Previously, the αH-αI loop was found to participate in substrate protein binding. A 33-amino acid stretch of the yeast PKA isoform TPK1 (amino acids 298–330), which includes the αH-αI loop, was sufficient to allow for substrate binding in a yeast two-hybrid assay (47). Moreover, evolutionarily conserved residues in the core of the kinase that link the active site with the C terminus mediate an elevated substrate-binding phenotype, when mutated to alanine, namely Tyr-208TPK1 (Tyr-164Cα), Glu-252TPK1 (Glu-208Cα), Asp-264TPK1 (Asp-220Cα), Trp-266TPK1 (Trp-222Cα), Arg-324TPK1 (Arg-280Cα), and Lys-336/His-338TPK1 (Lys-292/His-294Cα). Mutation of residues Glu-252TPK1, Arg-324TPK1, and Lys-336/His-338TPK1 resulted in inactivity of the kinase and prohibited binding to the yeast R subunit (BCY1) (47, 48), which is a substrate for autophosphorylation (49).
In our analysis of human Cα, we provide strong biochemical evidence for a role of the αH-αI loop Leu-277Cα in stabilizing RII subunit interaction and controlling cAMP-mediated holoenzyme activation. By means of molecular dynamics simulation, we propose that mutation of Leu-277Cα to Arg destabilizes the interaction network composed of Asp-271bRIIα, Arg-245bRIIα, and Arg-194Cα (Fig. 5), thus influencing the important interaction of the R subunit αA-helix with the activation loop (4). PKA activation of the mutant holoenzymes occurred at much lower cAMP concentrations (Table 2), which can be explained by the fact that Arg-245bRIIα (Arg-241bRIa) is involved in cAMP binding via Tyr-209bRIIα (Glu-200bRIα) (4, 50). Finally, Leu-277Cα could be involved in stabilizing the Arg-245/Asp-271bRIIα (Arg-241/Asp-267bRIα) salt bridge, which breaks upon cAMP binding, followed by the major collapse of the αB/C helix and subsequent holoenzyme dissociation (3, 4, 51). In summary, our work supports a critical involvement of the αH-αI loop in autoregulation of PrKX-like kinases (Arg-283PrKX), in stabilizing PKA type II holoenzymes (Leu-277Cα), and in proper allosteric signal propagation in PKA (Leu-277Cα), besides the previously recognized role of this region in substrate recognition (47, 48) and in stabilizing the active conformation of the kinase via the universally conserved Arg-280Cα residue (40, 52).
Supplementary Material
Acknowledgment
We acknowledge the help of Dr. Martina Rex with phylogenetic analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AA18060 from the National Science Foundation-NIH Collaborative Research in Computational Neuroscience Program (to M. Z.), European Union Grant LSHB-CT-2006-037189 (to F. W. H. and M. Z.), Bundesministerium für Bildung und Forschung NGFN2 Grant FKZ01GR0441 (to F. W. H.), and Deutsche Forschungsgemeinschaft BO1100/2 (to M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
M. Boshart, unpublished data.
M. Diskar and A. Prinz, unpublished data.
- C
- PKA catalytic subunit
- BRET
- bioluminescence resonance energy transfer
- IBMX
- 3-isobutyl-1-methylxanthine
- MD
- molecular dynamics
- PKA
- cAMP-dependent protein kinase A
- R
- PKA regulatory subunit
- Rluc
- Renilla luciferase
- PrKX
- protein kinase X
- PrKY
- protein kinase Y
- SPR
- surface plasmon resonance
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1.Herberg F. W., Taylor S. S., Dostmann W. R. (1996) Biochemistry 35, 2934–2942 [DOI] [PubMed] [Google Scholar]
- 2.Vigil D., Blumenthal D. K., Taylor S. S., Trewhella J. (2006) J. Mol. Biol. 357, 880–889 [DOI] [PubMed] [Google Scholar]
- 3.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 4.Wu J., Brown S. H., von Daake S., Taylor S. S. (2007) Science 318, 274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S. H., Wu J., Kim C., Alberto K., Taylor S. S. (2009) J. Mol. Biol. 393, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiebel K., Winkelmann M., Mertz A., Xu X., Page D. C., Weil D., Petit C., Rappold G. A. (1997) Hum. Mol. Genet. 6, 1985–1989 [DOI] [PubMed] [Google Scholar]
- 7.Skalhegg B. S., Tasken K. (2000) Front. Biosci. 5, D678–693 [DOI] [PubMed] [Google Scholar]
- 8.Li X., Li H. P., Amsler K., Hyink D., Wilson P. D., Burrow C. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9260–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junttila I., Bourette R. P., Rohrschneider L. R., Silvennoinen O. (2003) J. Leukocyte Biol. 73, 281–288 [DOI] [PubMed] [Google Scholar]
- 10.Semizarov D., Glesne D., Laouar A., Schiebel K., Huberman E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Burrow C. R., Polgar K., Hyink D. P., Gusella G. L., Wilson P. D. (2008) Biochim. Biophys. Acta 1782, 1–9 [DOI] [PubMed] [Google Scholar]
- 12.Li X., Hyink D. P., Polgar K., Gusella G. L., Wilson P. D., Burrow C. R. (2005) J. Am. Soc. Nephrol. 16, 3543–3552 [DOI] [PubMed] [Google Scholar]
- 13.Li X., Hyink D. P., Radbill B., Sudol M., Zhang H., Zheleznova N. N., Wilson P. D. (2009) Kidney Int. 76, 54–62 [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann B., Chiorini J. A., Ma Y., Kotin R. M., Herberg F. W. (1999) J. Biol. Chem. 274, 5370–5378 [DOI] [PubMed] [Google Scholar]
- 15.Diskar M., Zenn H. M., Kaupisch A., Prinz A., Herberg F. W. (2007) Cell. Signal. 19, 2024–2034 [DOI] [PubMed] [Google Scholar]
- 16.Prinz A., Diskar M., Erlbruch A., Herberg F. W. (2006) Cell. Signal. 18, 1616–1625 [DOI] [PubMed] [Google Scholar]
- 17.Prinz A., Diskar M., Herberg F. W. (2006) ChemBioChem 7, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 18.Gesellchen F., Prinz A., Zimmermann B., Herberg F. W. (2006) Eur. J. Cell Biol. 85, 663–672 [DOI] [PubMed] [Google Scholar]
- 19.Bertinetti D., Schweinsberg S., Hanke S. E., Schwede F., Bertinetti O., Drewianka S., Genieser H. G., Herberg F. W. (2009) BMC Chem. Biol. 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook P. F., Neville M. E., Jr., Vrana K. E., Hartl F. T., Roskoski R., Jr. (1982) Biochemistry 21, 5794–5799 [DOI] [PubMed] [Google Scholar]
- 21.Berrera M., Pantano S., Carloni P. (2006) Biophys. J. 90, 3428–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrera M., Pantano S., Carloni P. (2007) J. Phys. Chem. B 111, 1496–1501 [DOI] [PubMed] [Google Scholar]
- 23.Lindahl E., Hess B., van der Spoel D. (2001) J. Mol. Model. 7, 306–317 [Google Scholar]
- 24.Ponder J. W., Case D. A. (2003) Adv. Protein Chem. 66, 27–85 [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen W., Chandrasekhar J., Madura J., Impey R., Klein M. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 26.Hess B., Bekker H., Berendsen H., Fraaije J. (1997) J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 27.Sagui C., Darden T. A. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 155–179 [DOI] [PubMed] [Google Scholar]
- 28.Berendsen H., Postma J., Vangunsteren W., Dinola A., Haak J. (1984) J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 29.Eisenhaber F., Lijnzaad P., Argos C., Sander C., Scharf M. (1995) J. Comput. Chem. 16, 273–284 [Google Scholar]
- 30.Berrera M., Cattaneo A., Carloni P. (2006) Biophys. J. 91, 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaschke R. J., Monaghan A. P., Bock D., Rappold G. A. (2000) Genomics 64, 187–194 [DOI] [PubMed] [Google Scholar]
- 32.Li W., Yu Z. X., Kotin R. M. (2005) J. Histochem. Cytochem. 53, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 33.Meléndez A., Li W., Kalderon D. (1995) Genetics 141, 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. (2005) Science 309, 416–42216020726 [Google Scholar]
- 35.Martin B. R., Deerinck T. J., Ellisman M. H., Taylor S. S., Tsien R. Y. (2007) Chem. Biol. 14, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 36.Viste K., Kopperud R. K., Christensen A. E., Døskeland S. O. (2005) J. Biol. Chem. 280, 13279–13284 [DOI] [PubMed] [Google Scholar]
- 37.Dalton G. D., Dewey W. L. (2006) Neuropeptides 40, 23–34 [DOI] [PubMed] [Google Scholar]
- 38.Hanks S. K. (2003) Genome Biol. 4, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Kennedy E. J., Wu J., Deal M. S., Pennypacker J., Ghosh G., Taylor S. S. (2009) J. Biol. Chem. 284, 6241–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson D. A., Akamine P., Radzio-Andzelm E., Madhusudan M., Taylor S. S. (2001) Chem. Rev. 101, 2243–2270 [DOI] [PubMed] [Google Scholar]
- 41.Gibbs C. S., Knighton D. R., Sowadski J. M., Taylor S. S., Zoller M. J. (1992) J. Biol. Chem. 267, 4806–4814 [PubMed] [Google Scholar]
- 42.Anjard C., Etchebehere L., Pinaud S., Véron M., Reymond C. D. (1993) Biochemistry 32, 9532–9538 [DOI] [PubMed] [Google Scholar]
- 43.Etchebehere L. C., Van Bemmelen M. X., Anjard C., Traincard F., Assemat K., Reymond C., Véron M. (1997) Eur. J. Biochem. 248, 820–826 [DOI] [PubMed] [Google Scholar]
- 44.Bettencourt da Cruz A., Wentzell J., Kretzschmar D. (2008) J. Neurosci. 28, 10885–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubin C. S., Erlichman J., Rosen O. M. (1972) J. Biol. Chem. 247, 36–44 [PubMed] [Google Scholar]
- 46.Anand G. S., Hotchko M., Brown S. H., Ten Eyck L. F., Komives E. A., Taylor S. S. (2007) J. Mol. Biol. 374, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deminoff S. J., Ramachandran V., Herman P. K. (2009) Genetics 182, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deminoff S. J., Howard S. C., Hester A., Warner S., Herman P. K. (2006) Genetics 173, 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. (1987) Mol. Cell Biol. 7, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symcox M. M., Cauthron R. D., Ogreid D., Steinberg R. A. (1994) J. Biol. Chem. 269, 23025–23031 [PubMed] [Google Scholar]
- 51.Gullingsrud J., Kim C., Taylor S. S., McCammon J. A. (2006) Structure 14, 141–149 [DOI] [PubMed] [Google Scholar]
- 52.Steichen J. M., Iyer G. H., Li S., Saldanha S. A., Deal M. S., Woods V. L., Jr., Taylor S. S. (2010) J. Biol. Chem. 285, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.