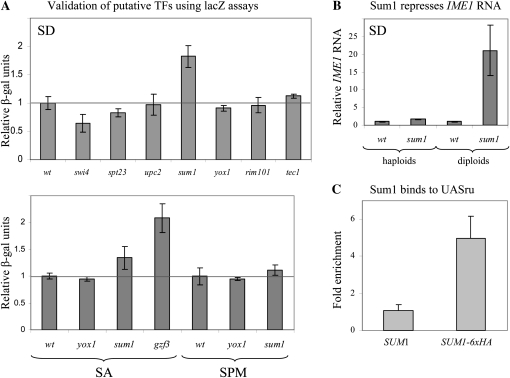
Figure 4.—
Validation of putative TFs identified by the R-SGA screen. (A) Samples were taken to extract proteins and measure lacZ levels following growth in either SD or SA media to 107 cells/ml. In addition, proteins were extracted from cells grown in SA media to 1 × 107 cells/ml and shifted to SPM for 6 hr. The relative level of β-gal is given. Strains used are wild-type (Y1214) and its isogenic swi4Δ (Y1855), spt23Δ (Y1860), upc2Δ (Y1859), sum1Δ (Y1827), yox1Δ (Y1824), rim101 (Y1854), and tec1Δ (Y1858) strains. These strains carry IME1UASru–HIS4TATA–lacZ integrated in the genomic LEU2 gene. Additional strains were the wild-type Y1825 and its isogenic gzf3Δ (Y1836). These strains carry IME1–lacZ integrated in the genomic LEU2 gene. The results are the averages of at least three independent colonies, and standard deviation is given. (B) RNA was isolated from cells grown in SD media to 1 × 107 cells/ml. Level of RNA was determined by qPCR. The relative level of IME1 RNA in comparison to RNA levels of ACT1 is given. Strains used are: Y1214 and Y1721, the wild-type haploid and diploid strains, respectively, and their isogenic sum1Δ (Y1827) and sum1Δ/sum1Δ (Y1891) strains. The results are the averages of at least three independent colonies. (C) Samples for ChIP assay were taken from 3 × 108 logarithmic cells grown in SD. Real-time PCR was used to amplify the specific UASru element present on IME1UASru–HIS4TATA–lacZ and the nonspecific TEL1. Fold enrichment of the specific vs. the nonspecific PCR products for IP without antibody (w/o α) or with antibody directed against the HA epitope (α HA) are given. The results are the averages of three independent colonies, and standard deviation is given. Strain used is Y1837.