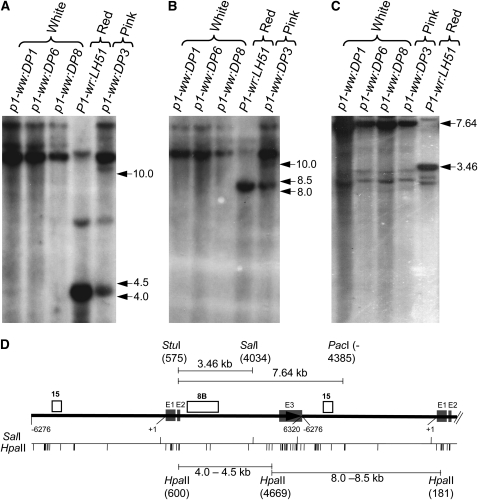
Figure 2.—
DNA gel-blot analysis showing that the silenced mutants are hypermethylated. Leaf genomic DNA of three white cob mutants, a pink cob mutant, and the red cob (wild-type) progenitor was used for the methylation assay. (A and B) Gel blot containing HpaII-digested DNA was hybridized with probe fragments 8B (A) and 15 (B). (C) DNA was simultaneously digested with a mixture of methylation-insensitive (StuI and PacI) and methylation-sensitive (SalI) enzymes, and the resulting blot was hybridized with probe 8B. Size (in kilobase pairs) and location of important hybridizing bands is to the right of each autoradiogram. (D) Gene structure and restriction map of P1-wr. Gene structure is based on Chopra et al. (1998), and the coordinates are consistent with GenBank accession EF165349 (Sekhon et al. 2007). The solid black arrow represents a complete P1-wr copy while the black solid line following the arrow represents a partial downstream copy. Shaded boxes (marked E1, E2, and E3) represent exons. Probes are shown above the gene structure diagram, and the coordinates are shown below. Also shown below the gene structure diagram are the positions of the HpaII and SalI sites. Some important restriction fragments, along with the location and coordinates of restriction enzyme sites yielding these fragments, are shown above and below the gene structure diagram.