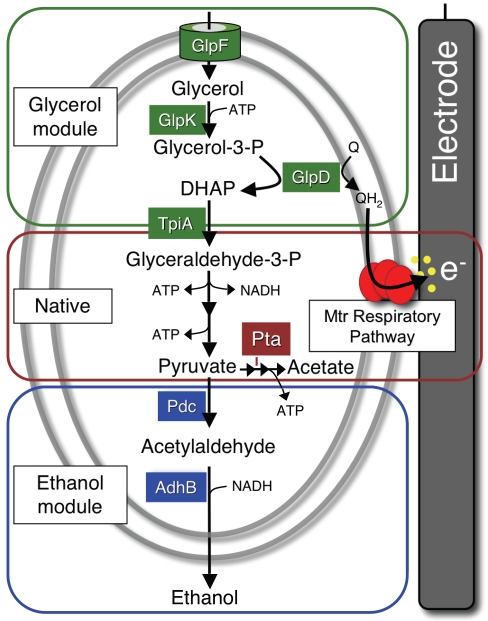
FIG 1 .
Metabolic modules added to S. oneidensis to enable electrode-dependent conversion of glycerol to ethanol. The glycerol utilization module from E. coli (section outlined in green [top]) and the ethanol production module from Z. mobilis (blue section [bottom]) were combined with native metabolic pathways (red section [middle]) for stoichiometric non-redox-balanced conversion of glycerol to ethanol. Metabolic substrates, intermediates, and products are denoted in black. The genes from E. coli and Z. mobilis engineered into S. oneidensis are represented by the proteins that they encode: GlpF, glycerol transporter; GlpK, glycerol kinase; GlpD, glycerol-3-phosphate dehydrogenase; Pdc, pyruvate decarboxylase; and AdhB, alcohol dehydrogenase. Phosphotransacetylase (Pta) is shown from native metabolism. Electrons not redox balanced within the cell are subsequently transferred through the Mtr pathway to the electrode. Yellow dots represent flavins secreted naturally from cells to accelerate extracellular transfer of electrons (e−) to the electrode.