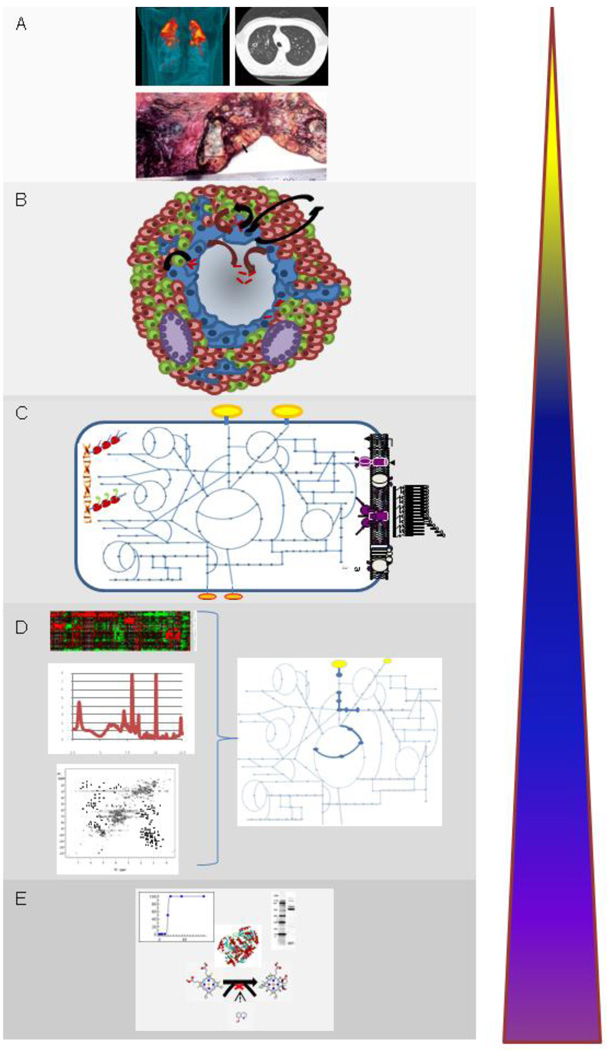
Figure 1.
Development of systems approaches for understanding MTb infection requires integrating information from all levels of understanding of disease. At the highest level (A), information is gathered from tuberculosis patients ranging from understanding the pathology of disease from clinical data and imaging studies and the epidemiology of human genetic variation in disease susceptibility and progression. Modeling of the dynamics of infection at the level of the granuloma (B) builds on information acquired from host immunology as well as pathogen biology. Genomic information of MTb as well as clinical strains of MTb has allowed metabolic networks (C) to be constructed that mirror the metabolic capabilities of this organism as well as the production of metabolites that play a role in growth, survival and virulence. Data gathered from “omics” studies such as transcriptomics, metabolomics and proteomics have allowed reconstruction of dynamic metabolic networks that capture information of metabolic fluxes that occur under various environmental conditions (D). The majority of work to date has been performed at the base level which includes focused studies of proteins, their function and structure and their interaction with small molecules (E).