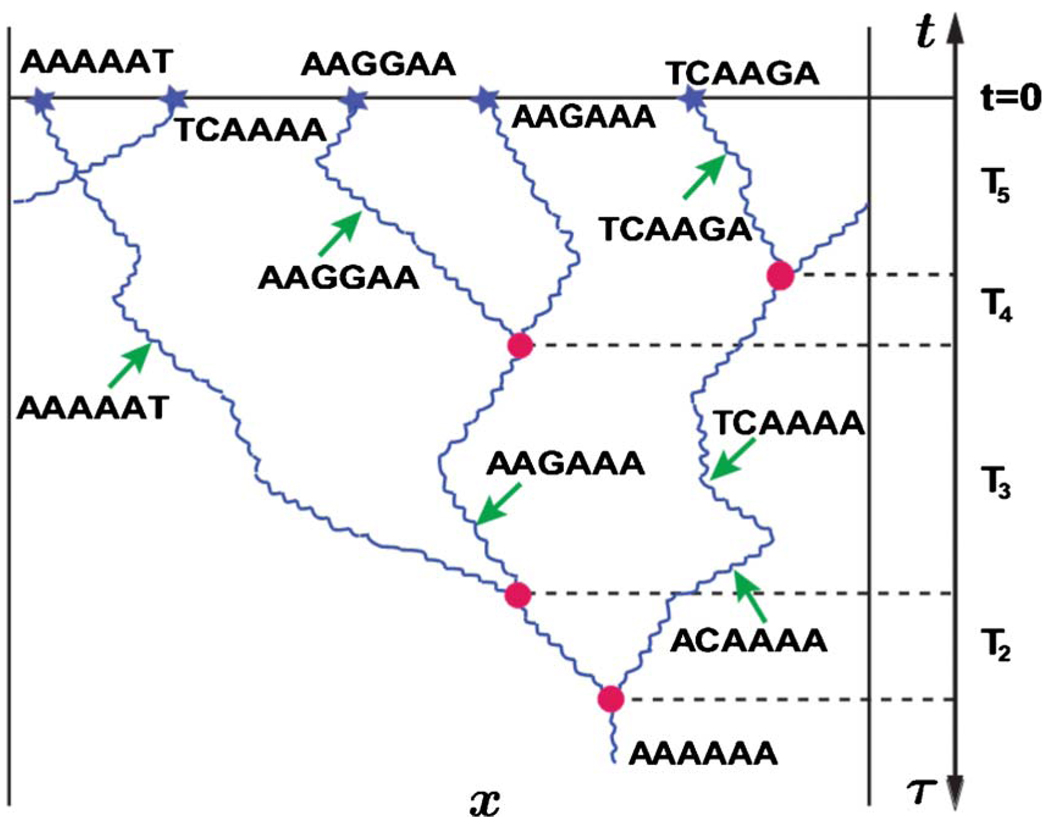
FIG. 17.
(Color online) An illustration of the backward-in-time dynamics of the ancestral lineages in a one-dimensional habitat with periodic boundary conditions. Five organisms (i.e., n=5) are sampled from the much larger population at t=0 and their DNA is sequenced. We do not display the sites that are identical for all organisms, which are usually the majority of the sequenced sites, i.e., only the segregating sites are shown. For illustration purposes, we also assumed that all samples differ in at least one nucleotide, but in experiments one often finds organisms that have identical sequences. We trace the spatial diffusion and coalescence of the lineages backward in time until they merge into a single lineage of the common ancestor of the whole sample. The coalescence events are denoted by circles and the mutations are denoted by arrows and the resulting mutated sequences. Note that lineages may cross without coalescing as shown in the top left corner. The ancestral process shown here satisfies the infinite site model and illustrates the fact that the more genetically similar the lineages are the more likely they are to have a common ancestor in the recent past. Most genetic inference methods rely on this relationship as shown in the text.