Figure 2.
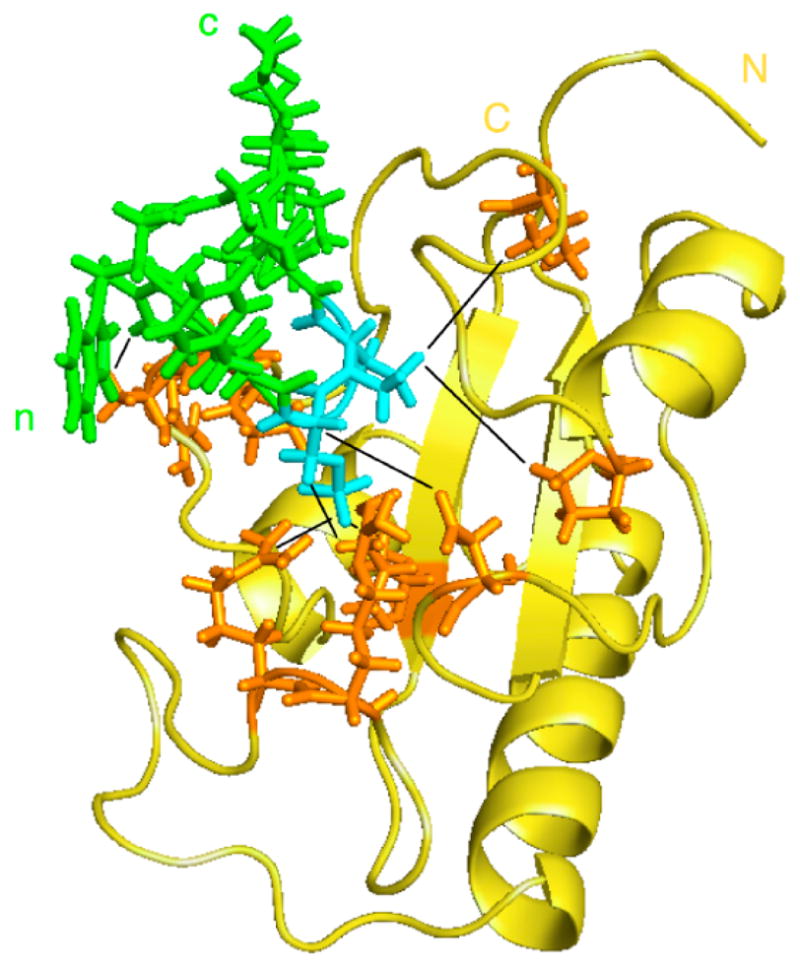
Model of the structure of the Pin1 catalytic domain in complex with the substrate peptide WFYpSPRLKK. Pin1cat is shown in gold with the sidechains of enzyme residues involved distance restraints depicted in orange. The substrate peptide is shown in green with the pS-Pro residues highlighted in cyan. The starting structure used was Pin1cat (PDB: 1PIN[37]) after removal of the Ala-Pro dipeptide and the sulfate ion in the active site. Distance constraints (shown as black lines) were defined based on intermolecular NOEs measured during catalytic turnover and on distances to the sulfate ion in the starting structure. Complex was energy-minimised and equilibrated using CHARMM[41].
