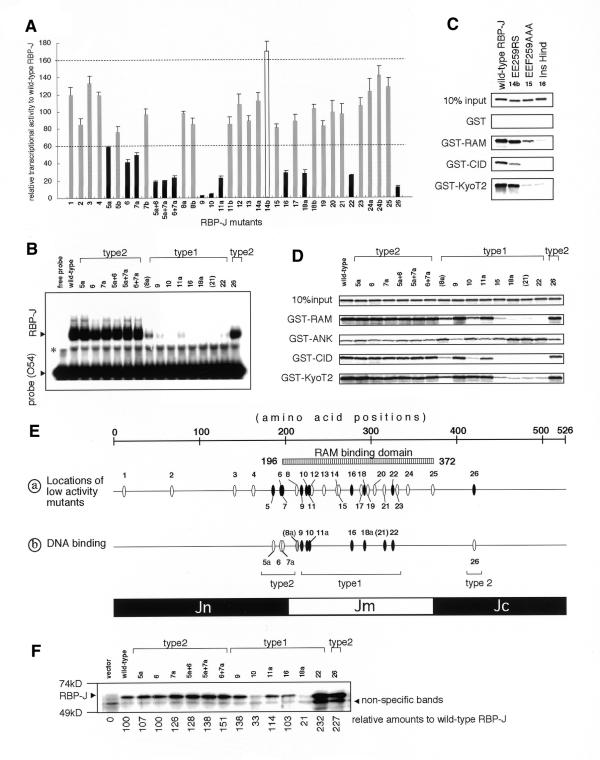
Figure 2.
(Opposite) Relative transcriptional activities of wild-type RBP-J and mutants by RAMIC and their relative binding activities for interacting molecules. RBP-J mutants are represented by numbers, as follow: 1, InsSpl; 2, InsDra; 3, InsPvu; 4, RK162GS-T153A; 5a, KR185GS; 5b, KR185RS; 6, S194A; 7a, KK196GS; 7b, KK196RS; 8a, KV212GS; 8b, KV212RS; 9, R218H; 10, InsKpn; 11a, RY227GS; 11b, RY227RS; 12, H230G; 13, GA244GS; 14a, EE259GS; 14b, EE259RS; 15, EEF259AAA; 16, InsHind; 17, RL287GS; 18a, RK291GS; 18b, RK291RS; 19, KQ295GS; 20, DD303GS; 21, FY314GS; 22, InsRsa971; 23, ER329GS; 24a, KE341GS; 24b, KE341RS; 25, InsApa; 26, InsRsa1265. The designations 5a+6, 5a+7a and 6+7a indicate the double mutants of KR185GS+S194A, KR185GS+KK196GS and S194A+KK196GS, respectively. (A) The peak transcriptional activities of RBP-J mutants relative to wild-type RBP-J. RBP-J mutants with high (>160%), normal (60–160%) and low (<60%) transcriptional activities are shown as open, shaded and closed bars, respectively. (B) EMSA of mutants with low transcriptional activity in addition to KV212GS (mutant 8a) and FY314GS (mutant 21) are shown. The same molar quantities of proteins were reacted with the 32P-labeled O54 probes containing the RBP-J binding sites. Designations of type 2 mutants are explained in the text. Since 8a and 21 are not type 1 mutants, they are parenthesized. The asterisk represents non-specific bands. (C) The GST pull-down assay of EE259RS (14b), a mutant with high transcriptional activity, for GST, GST–RAM, GST–CID and GST–KyoT2 are displayed in comparison with wild-type RBP-J, EEF259AAA (15), and InsHind (mutant 16, a negative control for the RAM-binding activity). (D) The GST pull-down assay of the mutants as in (C) for GST–RAM, GST–ANK, GST–CID and GST–KyoT2 are shown, including 10% input proteins. The remaining mutants are not shown in the GST pull-down assays or EMSAs because they revealed activity very similar to wild-type RBP-J. (E) Schematic summation of the results in (A), (C) and (D). (a) The positions of mutations aligned along RBP-J with reduced transcriptional activity mutants as shown in (A). Closed ovals represent mutants with decreased (<60%) transcriptional activities. Types 1 and 2 are explained in the text. (b) DNA-binding activities of RBP-J mutants analyzed in (C) are shown. Closed, shaded and open ovals show significantly decreased (<20% of wild-type RBP-J), slightly decreased (20–50%) and normal affinities, respectively. (F) Expression levels of mutants with low transcriptional activity in OT11 cells are shown. An aliquot of 1 µg of pCDM8 RBP-J and mutants was transfected in a 3.5 cm dish of OT11 cells and one-tenth of lysate was applied, blotted and detected by anti-RBP-J antibody (T6719). Detected results were adjusted by background amounts and shown as relative amounts to wild-type RBP-J (with wild-type set at 100). Although anti-RBP-J antibody displays lower affinities for mutants 10 and 18a (33 and unpublished data), significant levels of all the type 2 mutants are expressed. All experiments were confirmed in triplicate.