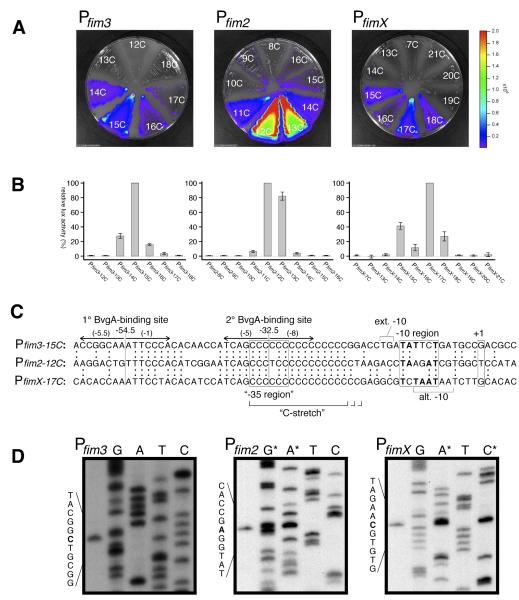
Fig. 1. fim promoter comparison. A&B) C-stretch optimization.
Variants of Pfim3, Pfim2, and PfimX that differed in the length of the C-stretch were constructed, cloned into the luxCDABE fusion vector pSS3967, and conjugated into B. pertussis strain BP536. Growth of the resulting chromosomal integrants and quantitation of light production is described in Materials and Methods. C) Alignment of optimal-length fim promoters. DNA sequences of fim promoters aligned by sequence identity, with identical nucleotides depicted by colons between juxtaposed sequences. Inverted arrows indicate the positions of BvgA-binding sites. Boxed regions correspond to the start of transcription (+1), the −10 region, and the expected position of a −35 element were one present (“−35 region”). Nucleotides in bold type within the boxed −10 regions correspond to those that match the consensus sequence TATAAT. Brackets indicate the positions of an extended −10 TG dinucleotide in the fim3 promoter, and an alternative −10 sequence in the fimX promoter. Numbers in parentheses above the inverted arrows indicate the predicted score for BvgA-binding half-sites in the fim3 promoter according to the algorithm represented in Fig. 2C. D) Transcription start site determination for fim3, fim2, and fimX promoters. Primer extension products and sequencing ladders are shown for each promoter. Sequencing ladders correspond to the sequence of the complementary (template) strand. In the fim2 and fimX panels, some of the sequence ladders (denoted by asterisk) were doped with primer extension product to unambiguously identify the starting nucleotide.