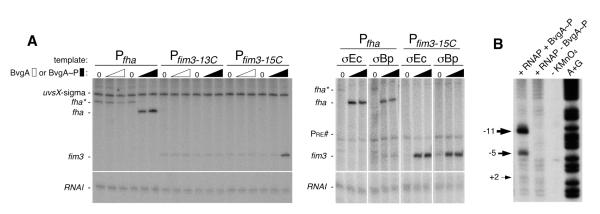
Fig. 3. BvgA~P activation of Pfim3 and Pfha and identification of Pfim3 transcription bubble in vitro.
A) Denaturing gels showing the products of in vitro transcription reactions, assembled as follows. Left panel: 0.25 pmol template DNA (0.2 pmol Pfha or Pfim3 plus 0.05 pmol PuvsX-sigma) was mixed with unphosphorylated or phosphorylated BvgA (0, 2, or 4 pmol) in 5.9 ul protein buffer I on ice. A solution containing 1.5 pmol of reconstituted RNAP in 4.1 ul protein buffer II was then added. Right panel: 0.1 pmol template DNA (0.05 pmol Pfha or Pfim3-15C plus 0.05 pmol Pre#) was mixed with phosphorylated BvgA (0, 1, 2pmol) in 5 ul protein buffer III on ice. A solution containing 0.75 pmol reconstituted RNAP in 5 ul protein buffer IV was then added. In each case, reactions were incubated for 15 min at 37° C, and transcription was initiated by adding 1 ul of NTP mix containing heparin to limit transcription to a single round. After incubation at 37° C for 10 min, reactions were collected on dry ice and treated as described in Materials and Methods. Sigma70-dependent promoters PRNAI (Morita & Oka, 1979), present on the supercoiled plasmid template, and Puvsx-sigma (Hinton & Vuthoori, 2000), added as a linear template, served as BvgA-independent controls. B) Denaturing gel showing the products of potassium permanganate footprinting of Pfim3 DNA that had been 32P-labeled at the 5′ end of the template (bottom) strand. The DNA was incubated with the indicated proteins with (lanes 1, 2) or without (lane 3) potassium permanganate or was used in an A+G sequencing reaction (lane 4) (Maxam & Gilbert, 1980) to allow determination of cleavage coordinates, numbered relative to the start of transcription.