Abstract
During female meiosis, bivalent chromosomes are thought to be held together from birth until ovulation by sister chromatid cohesion mediated by cohesin complexes whose ring structure depends on kleisin subunits, either Rec8 or Scc1. Because cohesion is established at DNA replication in the embryo, its maintenance for such a long time may require cohesin turnover. To address whether Rec8- or Scc1-containing cohesin holds bivalents together and whether it turns over, we created mice whose kleisin subunits can be cleaved by TEV protease. We show by microinjection experiments and confocal live-cell imaging that Rec8 cleavage triggers chiasmata resolution during meiosis I and sister centromere disjunction during meiosis II, while Scc1 cleavage triggers sister chromatid disjunction in the first embryonic mitosis, demonstrating a dramatic transition from Rec8- to Scc1-containing cohesin at fertilization. Crucially, activation of an ectopic Rec8 transgene during the growing phase of Rec8TEV/TEV oocytes does not prevent TEV-mediated bivalent destruction, implying little or no cohesin turnover for ≥2 wk during oocyte growth. We suggest that the inability of oocytes to regenerate cohesion may contribute to age-related meiosis I errors.
Keywords: Cohesin, Rec8, meiosis, oocytes
During meiosis, two rounds of chromosome segregation without an intervening round of DNA replication generate haploid gametes from diploid germ cells. Following meiotic DNA replication, reciprocal recombination between nonsister homologous chromatids creates bivalent chromosomes whose four DNA molecules are thought to be held together by cohesion between sister chromatids initially established during S phase. A bundling of sister centromeres into a single kinetochore ensures that maternal and paternal centromeres are pulled in opposite directions by meiosis I microtubules, a state stabilized by tension (Nicklas 1997) made possible by cohesion distal to the crossover sites. Meiosis I chromosome segregation is accompanied by the selective loss of cohesion along chromosome arms and the disjunction to opposite poles of dyad chromosomes containing a parental and a recombinant chromatid attached to each other by cohesion persisting at centromeres. Dyads are subsequently converted to individual chromatids during the second meiotic division (Petronczki et al. 2003).
A remarkable and still mysterious feature of meiosis in the germline of most female vertebrates is a prolonged prophase arrest following formation of bivalent chromosomes, which lasts from around birth until ovulation, an interval that might be many months in the mouse but several decades in humans. Before ovulation and completion of the first meiotic division, oocytes must first undergo an extensive hormone-dependent growth phase that lasts 85 d in humans and ≥14 d in mice. It is suspected that it is during their extended prophase arrest that oocytes age in a manner that greatly increases the frequency of chromosome missegregation at both meiotic divisions, a phenomenon largely responsible for age-related infertility and trisomy. The relationship between maternal age and trisomy is one of the most important etiological factors associated with any human genetic disorder (Hassold and Hunt 2001; Hassold et al. 2007).
What goes so badly awry as oocytes age? One candidate is the cohesion between sister chromatids necessary to hold bivalent chromosomes together. Work in yeast has shown that sister chromatid cohesion in mitotic cells is mediated by a multisubunit complex called cohesin whose Smc1, Smc3, and α-kleisin subunits form a huge tripartite ring within which it is proposed that sister DNAs are entrapped during the process of DNA replication (Nasmyth and Haering 2009). Cleavage of cohesin's kleisin subunit by a thiol protease called separase, activated only after all chromosomes have bioriented on spindles, opens the ring, thereby permitting traction of sister DNAs to opposite poles at the metaphase-to-anaphase transition. The findings that cohesion is established during DNA replication in yeast (Uhlmann and Nasmyth 1998), that it cannot normally be regenerated thereafter (Haering et al. 2004), and that cohesin is also responsible for holding bivalents together (Buonomo et al. 2000) have raised the possibility that it might be the Achilles' heal of ageing oocytes. According to this hypothesis, age-related cohesin decay along chromosome arms either causes the terminalization and eventual loss of chiasmata, compromising segregation during meiosis I, or leads to insufficient cohesion persisting after meiosis I at centromeres, compromising segregation during meiosis II. Consistent with this hypothesis is the finding that precocious sister chromatid disjunction greatly increases in aging oocytes whose cohesion has been compromised by inactivation of a Smc1 isoform (Hodges et al. 2005).
The cohesin decay hypothesis is consistent with the finding that trisomy 21 arising from meiosis I errors is often associated with distal crossovers, while trisomy arising from meiosis II errors is associated with crossing over close to centromeres, which would further weaken centromeric cohesion (Oliver et al. 2008; Ghosh et al. 2009). However, the recent finding that cohesin can build new cohesion throughout the genome of post-replicative yeast cells with damaged DNA (Ström et al. 2007; Unal et al. 2007) has raised the prospect that chromosome segregation in a 45-yr-old woman might not, after all, depend on 45-yr-old cohesin proteins. This notion has been buttressed by the report that Smc1β, a meiosis-specific variant of Smc1 that helps to hold bivalents together in mouse oocytes, only associates with chromosomes after DNA replication, and low but detectable levels of mRNA persist in oocytes of sexually mature females (Hodges et al. 2005).
Whether cohesin can continue to build new bridges that knit bivalents together long after chiasmata are formed is a question whose answer may be pivotal in understanding the etiology of age-related aneuploidy. To address it, we set out to determine the identity of the cohesin complexes that hold bivalent and dyad chromosomes together. Mammals possess two genes encoding α-kleisin proteins: Scc1/Rad21 is expressed in both mitotic and meiotic cells, and a distant relative called Rec8 is expressed in spermatocytes and oocytes but in few if any somatic tissues other than the thymus (Lee et al. 2002). Based on immunolocalization studies, it has variously been proposed that Scc1 participates in meiotic sister chromatid cohesion (Parra et al. 2004; Prieto et al. 2004; Xu et al. 2004, 2005) along with Rec8 (Lee et al. 2002; Eijpe et al. 2003) and/or that it promotes mono-orientation of sister kinetochores in meiosis I (Parra et al. 2004; Gomez et al. 2007). The supposed lack of either Rec8 or Scc1 on metaphase II chromosomes has even led to the suggestion that the cohesion between sister centromeres holding dyads together is mediated by a cohesin-independent mechanism (Parra et al. 2004; Gomez et al. 2007). Although subsequent studies have detected Rec8, but not Scc1, on metaphase II centromeres (Eijpe et al. 2003; Lee et al. 2003; Kudo et al. 2006, 2009) and shown that its persistence after meiosis I depends on MeiS332/shugoshin-like proteins that prevent the precocious loss of centromere cohesion (Llano et al. 2008), there is no direct demonstration that Rec8-containing cohesin complexes hold dyads together in any eukaryotic organism.
Our ignorance concerning the identity of cohesin subunits mediating cohesion in mammalian meiotic cells stems from the fact that Smc1, Smc3, and Scc1 are essential for cell proliferation, which naturally hinders analysis of meiotic functions, while spermatocytes and oocytes lacking Rec8 fail to progress beyond pachytene (Bannister et al. 2004; Xu et al. 2005), presumably due to defects in double-strand break repair. In yeast, analyses of Rec8 alleles that cannot be cleaved by separase have been highly informative as to its role in chromosome segregation (Buonomo et al. 2000; Kitajima et al. 2003), but this approach has been less successful in animal cells. Males expressing a Rec8 variant that cannot be cleaved by separase in vitro do not complete meiosis I and are infertile, while females are fertile (Kudo et al. 2009). Both male and female germ cells manage eventually to resolve chiasmata, possibly due to persistent albeit inefficient Rec8 cleavage in vivo, but conceivably due to participation of another α-kleisin subunit.
We describe here a very different approach to investigating cohesin function in oocytes; namely, the use of homologous recombination to create mice whose Rec8 and Scc1 proteins contain cleavage sites for the TEV protease, a method that permits rapid inactivation of pre-existing cohesin complexes (Uhlmann et al. 2000; Oliveira et al. 2010). Microinjection experiments demonstrate that there is a dramatic switch from Rec8- to Scc1-containing cohesin complexes at the oocyte-to-zygote transition. Sister chromatids within zygotes (fertilized eggs) are held together exclusively by Scc1-cohesin. Prior to fertilization, in contrast, bivalent and dyad chromosomes are held together exclusively by Rec8-cohesin, which turns over little or not at all during the growing phase of oocytes.
Results
A TEV protease cleavage system to inactivate cohesin in mice
To address the identity of the kleisin subunit of cohesin complexes holding together meiotic chromosomes in mice, we replaced Rec8 and Scc1 genes with versions that express proteins containing TEV cleavage sites within the flexible polypeptide connecting the N-terminal and C-terminal domains that bind Smc3 and Smc1 ATP nucleotide-binding domains, respectively (Fig. 1A). Proteolytic cleavage using TEV mimics a natural process known to destroy sister chromatid cohesion in yeast; namely, cleavage of these subunits by separase at the metaphase-to-anaphase transition. Crucially, it enables inactivation even at late stages of meiosis of cohesin complexes that have already established sister chromatid cohesion, which cannot be achieved by gene knockouts or RNA knockdown. In both yeast and flies, TEV cleavage of Scc1/Rad21 in mitotic cells severs the cohesin ring, releases it from chromosomes, and abolishes sister chromatid cohesion within minutes (Uhlmann et al. 2000; Oliveira et al. 2010).
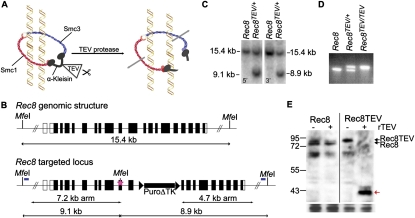
Figure 1.
Generation of a TEV-cleavable Rec8 allele. (A) Schematic of the cohesin ring consisting of an Smc1/Smc3 heterodimer bridged by the α-kleisin subunit. TEV protease-mediated cleavage of an α-kleisin subunit engineered to contain TEV protease recognition sites allows inducible opening of the ring. (B) Targeting strategy for generating a Rec8TEV allele. Schematic of the Rec8 genomic locus and targeted allele (Rec8TEV). Translated exons are shown as filled boxes. The pink star indicates three tandem TEV protease recognition sites, including a novel MfeI site, that were inserted into exon 10. The puromycin-truncated thymidine kinase selection cassette (rectangle) is flanked by LoxP sites (triangles). The relative location of the 5′ and 3′ probes (blue bars) and MfeI sites that were used in Southern blot analysis are shown on the targeted locus. (C) Southern blot analysis of wild-type (Rec8) and heterozygous (Rec8TEV/+) mouse ES genomic DNA (gDNA) digested with MfeI and hybridized with 5′ and 3′ probes to check for homologous recombination. (D) PCR analysis of wild-type (Rec8), heterozygous (Rec8TEV/+), and homozygous (Rec8TEV/TEV) mouse ear gDNA. (E) Western blot analysis of Rec8 cleaved by recombinant TEV protease (rTEV) in testes chromatin extracts from wild-type and Rec8TEV/TEV mice. Rec8 was detected by anti-Rec8 antibody. The red arrow indicates the Rec8 cleavage fragment.
We used gene targeting in mouse embryonic stem (ES) cells to insert a tandem array of three TEV protease recognition sequences into two sites each of Rec8 and Scc1 (Supplemental Figs. 1, 2), leaving known or putative separase cleavage sites intact. Southern blotting verified the presence of TEV recognition sequences containing a novel restriction site in either exon 10 (Fig. 1B,C) or exon 15 (Supplemental Fig. 1) of Rec8. Mice were generated using standard methods and genotyped by Southern blotting and PCR (Fig. 1D). To delete the selection cassette flanked by LoxP sites, mice were crossed to a strain expressing Cre recombinase under the control of the Sox2 promoter (Hayashi et al. 2002). Heterozygous Rec8e10TEV/+ intercrosses produced fully fertile Rec8e10TEV/e10TEV offspring with an average litter size of 7.2 pups (166 deliveries from 36 breeding pairs). Heterozygous Rec8e15TEVMyc/+ intercrosses produced infertile Rec8e15TEVMyc/e15TEVMyc offspring, indicating that this allele is nonfunctional. As described below, Rec8-Myc protein is functional because mice in which the sole source of Rec8 is a BAC transgene expressing a version containing nine tandem c-Myc epitopes at its C terminus are fertile (Kudo et al. 2006). This suggests that insertion of TEV sites into exon 15 inactivates Rec8 rather than the C-terminal c-Myc epitope tag. All experiments were therefore performed using the functional Rec8e10TEV allele (Fig. 1B), which will hereafter be referred to as Rec8TEV. Due to the paucity of material, it is difficult to demonstrate TEV-induced Rec8 cleavage in oocytes. We therefore tested whether the protease cleaves Rec8 in chromatin prepared from testes. TEV abolished full-length Rec8 and generated a cleavage fragment in Rec8TEV/TEV but not Rec8 extracts (Fig. 1E).
To compare the contributions of Rec8 and Scc1, we inserted TEV sites into Scc1. Southern blotting verified the presence of TEV sites within exon 11 and insertion of a C-terminal c-Myc10 epitope tag, both containing novel restriction sites (Fig. 2A,B). ES clones containing the c-Myc tag without the TEV site insertions were also identified (Fig. 2B). The c-Myc tag was added to compare Rec8-Myc and Scc1-Myc localization on chromosomes using anti-c-Myc antibody. An additional allele was generated by inserting TEV sites into exon 8 (Supplemental Fig. 2A). To test for functionality of the Scc1e11TEVMyc, Scc1Myc, and Scc1e8TEV alleles, the selection cassette flanked by LoxP sites was removed by transient Cre expression and a second round of targeting was performed using the original vector with the aim of generating homozygous cells. Double-targeted clones were identified for Scc1e11TEVMyc/e11TEVMyc and Scc1Myc/Myc but not Scc1e8TEV, demonstrating that Scc1e11TEVMyc and Scc1Myc alleles are functional in ES cells. All experiments described here were performed with the Scc1e11TEVMyc allele, which will hereafter be referred to as Scc1TEVMyc.
Figure 2.
Generation of a TEV-cleavable Scc1 allele. (A) Targeting strategy for generating an Scc1TEVMyc allele. Schematic of the Scc1 genomic locus and targeted alleles (Scc1TEVMyc or Scc1Myc). The pink star indicates three tandem TEV protease recognition sites, including a novel BamHI restriction site, that were inserted into exon 11. The orange star indicates a c-Myc10 epitope, including a novel BamHI restriction site that was inserted before the Stop codon. The relative location of the 5′ and 3′ probes (blue bars) and BamHI sites that were used in Southern blot analysis are shown on the targeted locus. (B) Southern blot analysis of wild-type (Scc1) and heterozygous (Scc1TEVMyc/+ and Scc1Myc/+) mouse ES gDNA digested with BamHI and hybridized with 5′ and 3′ probes to check for homologous recombination. (C) PCR analysis of wild-type (Scc1), heterozygous (Scc1TEVMyc/+), and homozygous (Scc1TEVMyc/TEVMyc) mouse ear gDNA (left panel), and wild-type (Scc1), heterozygous (Scc1Myc/+), and homozygous (Scc1Myc/Myc) mouse ear gDNA (right panel). (D) Western blot analysis of Scc1 cleavage by recombinant TEV protease in soluble and chromatin extracts from Scc1Myc/Myc, Scc1Myc/TEVMyc, and Scc1TEVMyc/TEVMyc ES cells. Scc1TEV-Myc and Scc1-Myc were detected by anti-c-Myc antibody. The red arrow indicates the Scc1 cleavage fragment.
Mice were generated by standard methods and targeted alleles of Scc1TEVMyc and Scc1Myc transmitted through the germline. Mice were genotyped by Southern blotting and PCR (Fig. 2B,C), and the selection cassette was deleted by crossing to a (Tg)Sox2-cre deleter strain. Heterozygous Scc1TEVMyc/+ intercrosses produced viable Scc1TEVMyc/TEVMyc offspring at sub-Mendelian ratios (Supplemental Fig. 2B). Similarly, heterozygous Scc1Myc/+ intercrosses produced viable Scc1Myc/Myc offspring at sub-Mendelian ratios (Supplemental Fig. 2B). Since the c-Myc tag is common to both alleles, it is possible that it affects an aspect of Scc1 function during embryonic development. However, since Scc1TEVMyc/TEVMyc and Scc1Myc/Myc mice are viable and produce offspring, we conclude that these modified Scc1 alleles are largely functional. To test whether TEV cleaves Scc1, we prepared extracts from ES cells. TEV abolished full-length soluble and chromatin-bound Scc1 and generated a cleavage fragment in Scc1TEV-Myc but not Scc1-Myc extracts (Fig. 2D).
TEV-induced Rec8 cleavage triggers chiasmata resolution
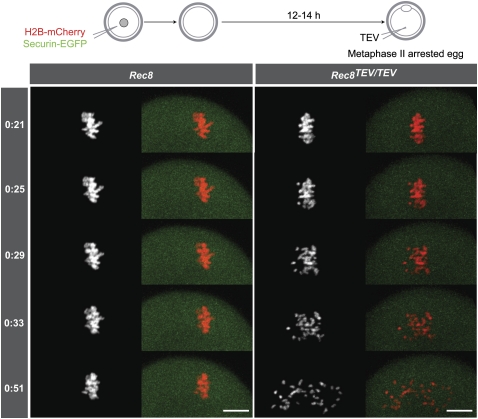
To image the first meiotic division in Rec8TEV/TEV females, fully grown germinal vesicle (GV)-stage oocytes were harvested in the presence of a phosphodiesterase inhibitor, 3-isobutyl-1-methylxanthine (IBMX), to maintain the GV arrest, and were microinjected with mRNA encoding histone H2B-mCherry to mark chromosomes and Securin-EGFP to measure APC/C activity (McGuinness et al. 2009). Upon removal of IBMX, GV breakdown (GVBD) is followed by alignment of bivalents on the metaphase I spindle. Their conversion to U-shaped dyad chromosomes (chiasmata resolution) occurs as Securin-EGFP fluorescence levels drop due to APC/C activation and is accompanied by chromosome segregation followed by polar body extrusion (PBE) in <1 h (Supplemental Fig. 3A), a course of events identical in Rec8 and Rec8TEV/TEV oocytes.
To test whether Rec8 cleavage is sufficient to trigger chiasmata resolution, we used a double-microinjection protocol. Wild-type or Rec8TEV/TEV GV oocytes were first injected with mRNA encoding H2B-mCherry, Securin-EGFP, and Flag-Mad2. By inhibiting cyclin B and securin degradation by the APC/C, the excess of Mad2 causes a metaphase I arrest (Wassmann et al. 2003). After culturing for 14 h post-GVBD, arrested oocytes were reinjected, this time with mRNA for TEV protease. TEV mRNA injection triggered in Rec8TEV/TEV (n = 5), but not in Rec8 oocytes (n = 7), conversion of bivalents first into dyads and subsequently into individual chromatids (Fig. 3A). TEV-induced Rec8 cleavage did not cause PBE for at least 4 h, presumably because cytokinesis requires cyclin-dependent kinase (CDK) inactivation. Importantly, Securin-EGFP levels remained high throughout, demonstrating that APC/C and separase had not been activated (Supplemental Fig. 4A,B). The resolution of chiasmata must therefore have been caused directly by TEV-induced cleavage. The chromatids produced by Rec8 cleavage in these experiments moved from pole to pole (Supplemental Movie 1).
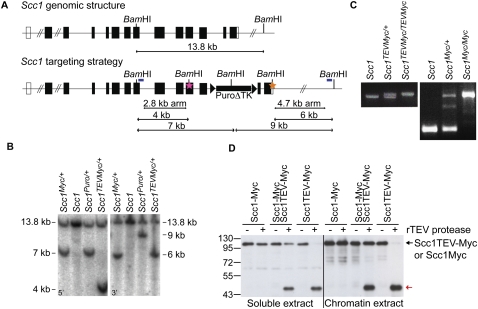
Figure 3.
Rec8 cleavage triggers chiasmata resolution and sister centromere disjunction of bivalent chromosomes. (A) Wild-type and Rec8TEV/TEV oocytes expressing H2B-mCherry, Securin-EGFP, and Flag-Mad2 were cultured for 14–16 h. Metaphase I-arrested oocytes were injected with TEV protease mRNA (time 0), and chromosome movements were visualized by time-lapse confocal microscopy (h:mm). Still images from a representative movie of each genotype are shown. The left panels show the H2B-mCherry channel and the right panels show H2B-mCherry and Securin-EGFP pseudocolored in red and green, respectively. Bar, 10 μm. (B) Wild-type and Rec8TEV/TEV oocytes expressing TEV protease were matured in vitro for 5 h. Chromosome spreads were prepared and stained with Hoechst to visualize DNA (red) and CREST to mark centromeres (green).
Chromosome spreads prepared 4–5 h post-GVBD from Rec8TEV/TEV oocytes that had been injected with TEV mRNA at the GV stage confirmed the presence of chromatids. The observed pattern suggests that TEV had indeed converted bivalents to chromatids. Meiosis I chromosome segregation normally occurs 6–8 h after GVBD. Rec8 TEV-injected and Rec8TEV/TEV-uninjected oocytes (n = 26) contained 20 bivalent chromosomes, while Rec8TEV/TEV oocytes (n = 18) injected with TEV contained only single chromatids (74–80 per cell) (Fig. 3B). Because similar experiments conducted with Scc1TEVMyc/TEVMyc oocytes revealed that meiotic chromosomes were unaffected by TEV-induced Scc1 cleavage (Supplemental Fig. 3B) and little or no Scc1 can be detected on bivalent chromosomes (Supplemental Fig. 3C), despite Scc1 being abundant in GVs (see Fig. 6B, below), we conclude that bivalent chromosomes are held together by cohesin containing Rec8. If we assume that this is engaged in holding sister chromatids together, then our experiments also demonstrate that it is sister chromatid cohesion and not some “chiasma binder” protein (Maguire 1974) that holds bivalents together.
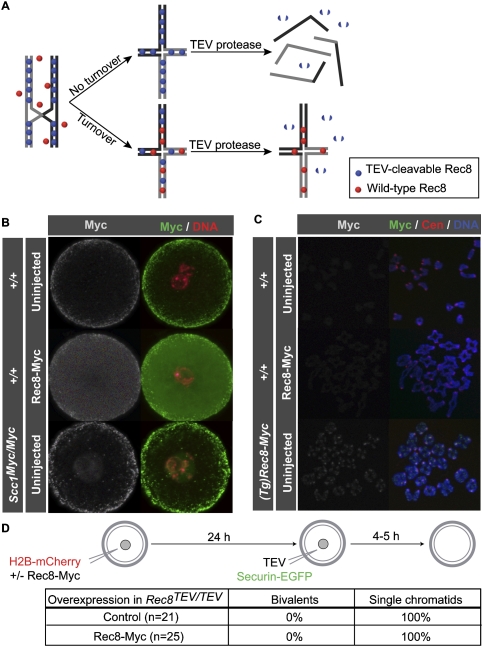
Figure 6.
No cohesin turnover in mature GV oocytes. (A) Schematic of cohesin turnover assay. TEV-cleavable Rec8 (blue spheres) is loaded during meiotic DNA replication and maintains the bivalent structure. If cohesin does not turn over following cohesion establishment and recombination, then wild-type Rec8 (red spheres) expressed post-recombination fails to be incorporated into functional cohesin complexes and TEV cleavage resolves bivalent chromosomes into single chromatids. If, on the other hand, cohesin turns over, then wild-type Rec8 expressed post-recombination forms de novo cohesion and the bivalent structure becomes resistant to TEV cleavage. (B) Wild-type GV oocytes were either injected with Rec8-Myc mRNA (middle panel) or uninjected (top panel) and cultured in the presence of IBMX for 24 h prior to fixation. (Bottom panel) An Scc1Myc/Myc GV oocyte was included as an uninjected control for c-Myc staining. Oocyte in situ immunostaining was performed using c-Myc antibody to visualize either overexpressed Rec8-Myc or endogenous Scc1-Myc (green), and DNA was stained with Hoechst (red). (C) Wild-type GV oocytes were either injected with Rec8-Myc mRNA (middle panel) or uninjected (top panel) and cultured in the presence of IBMX for 24 h. Oocytes were then cultured in IBMX-free medium for 5 h. Chromosome spreads were prepared and stained with c-Myc antibody to visualize either overexpressed Rec8-Myc or Rec8-Myc expressed from a constitutive BAC transgene, (Tg)Rec8-Myc (green), CREST to mark centromeres (red), and Hoechst to visualize DNA (red). (D) Rec8TEV/TEV oocytes expressing H2B-mCherry with or without Rec8-Myc were cultured in the presence of IBMX for 24 h and then injected with TEV protease and Securin-EGFP mRNA. Oocytes were released into IBMX-free culture medium and chromosomes were visualized by time-lapse confocal microscopy. Oocytes were scored as containing either 20 bivalent chromosomes or at least 72 single chromatids (and no bivalents) by 5 h post-GVBD.
TEV-induced Rec8 cleavage causes sister centromere disjunction in meiosis II
The special nature of meiotic centromeric cohesion has long been recognized. To explain its ability to survive after meiosis I, it was suggested that late replication of centromeric DNAs holds dyads together (Murray and Szostak 1985). The discovery that MeiS332/shugoshin-like proteins prevent loss of sister chromatid cohesion in the vicinity of centromeres but not along chromosome arms at the first meiotic division also raised the possibility that centromeric cohesion might differ from that along arms (Miyazaki and Orr-Weaver 1994), a notion supported by reports that cohesin is not readily detected at meiosis II centromeres in mammals (Parra et al. 2004; Gomez et al. 2007). However, the subsequent finding that Rec8-containing cohesin complexes persist at centromeres (Eijpe et al. 2003; Lee et al. 2003; Kudo et al. 2006) in a manner dependent on shugoshins (Lee et al. 2008) suggests that arm and centromere cohesion may, after all, be mediated by the same cohesin complex, whose cleavage by separase depends on phosphorylation of its Rec8 subunit and is prevented at centromeres during meiosis I by their recruitment of PP2A bound to shugoshins (Ishiguro et al. 2010; Katis et al. 2010).
Our creation of oocytes with TEV-cleavable kleisins enabled us to address the nature of meiotic centromeric cohesion directly. If Rec8-containing cohesin complexes also hold sister centromeres together, then TEV should induce sister centromere disjunction in Rec8TEV/TEV oocytes arrested in meiosis II by cystostatic factor (CSF). To test this, GV oocytes were injected with mRNA for H2B-mCherry and Securin-EGFP and allowed to undergo PBE in culture, and naturally arrested metaphase II eggs were injected with mRNA encoding TEV protease. TEV induced sister centromere disjunction in Rec8TEV/TEV (n = 8) but not Rec8 eggs (n = 7) (Fig. 4; Supplemental Movie 2). Importantly, Rec8 cleavage did not trigger a second PBE and Securin-EGFP levels remained high, demonstrating that both the APC/C and separase remained inactive. Similar experiments carried out with Scc1TEVMyc/TEVMyc eggs had no effect on sister centromere cohesion (Supplemental Fig. 5). We conclude that Rec8-containing cohesin complexes are solely responsible for chromosome arm and centromere cohesion in meiosis.
Figure 4.
Rec8 cleavage triggers sister centromere disjunction in meiosis II. Wild-type and Rec8TEV/TEV oocytes expressing H2B-mCherry and Securin-EGFP were cultured for 12–14 h. Metaphase II-arrested eggs were injected with TEV protease (time 0), and chromosome movements were visualized by time-lapse confocal microscopy (h:mm). Bar, 10 μm.
Fertilization triggers a switch from Rec8- to Scc1-containing cohesin
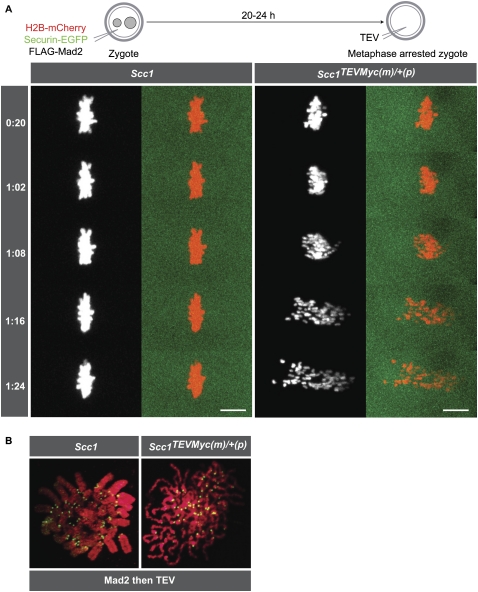
Rec8-cohesin maintains sister chromatid cohesion in eggs while Scc1-cohesin maintains cohesion in mitotically dividing cells, at least in yeast and fruit flies (Uhlmann et al. 2000; Vass et al. 2003; Pauli et al. 2008; Oliveira et al. 2010). There must therefore be a transition from meiotic to mitotic α-kleisin subunits at some point following fertilization. To address whether this is a gradual process taking place during early embryonic development or occurs suddenly in one step at the oocyte-to-zygote transition, Scc1TEVMyc/TEVMyc females were time-mated to wild-type males, rather than Scc1TEVMyc/TEVMyc males, which are not reliable breeders. This experimental setup has the potential caveat that zygotes will inherit an Scc1 allele from the male, resulting in genetically heterozygous Scc1TEVMyc(m)/+(p) embryos (m and p denote maternal and paternal alleles, respectively). However, this should not be a problem, as there is supposedly little or no essential transcription during the first zygotic cell cycle (Johnson 1981; Schultz 2002) and maternal Scc1TEVMyc would be expected to be the source of Scc1 in our zygotes. Consistent with this, we can only detect Scc1-Myc in zygotes produced by mating Scc1Myc/Myc females with wild-type males and not in zygotes produced by mating wild-type females with Scc1Myc/Myc males (Supplemental Fig. 6A).
To avoid interfering with cohesion establishment or other cohesin functions during the zygote's first cell cycle, pronuclear stage zygotes (G1/S phase, 6–10 h post-fertilization) were injected with H2B-mCherry, Securin-EGFP, and Flag-Mad2 mRNA to arrest them in metaphase of the first cell cycle, which normally lasts ∼24 h (from fertilization to cleavage). After expression for 20–24 h, arrested zygotes were reinjected with TEV protease mRNA. TEV mRNA injection triggered in Scc1TEVMyc(m)/+(p) (n = 9), but not in wild-type zygotes (n = 7), conversion of sister chromatids into single chromatids that subsequently moved from pole to pole (Fig. 5A; Supplemental Movie 3). The onset of sister chromatid disjunction following TEV protease injection was slower in zygotes than in eggs, which might reflect Scc1's greater abundance. TEV-induced Scc1 cleavage did not trigger cell division and Securin-EGFP levels remained high throughout, demonstrating that the APC/C and separase had not been activated. Sister chromatid disjunction must therefore have been caused directly by TEV-induced cleavage of Scc1.
Figure 5.
Scc1 cleavage triggers sister chromatid disjunction in the first embryonic mitosis. (A) Wild-type and Scc1TEVMyc/TEVMyc females were mated to wild-type males, and wild-type and Scc1TEVMyc(m)/+(p) zygotes (maternal vs. paternal alleles) were isolated, respectively. Zygotes expressing H2B-mCherry, Securin-EGFP, and Flag-Mad2 were cultured for 20–24 h. Metaphase-arrested zygotes were injected with TEV protease mRNA (time 0), and chromosome movements were visualized by time-lapse confocal microscopy (h:mm). Bar, 10 μm. (B) Wild-type and Scc1TEVMyc(m)/+(p) zygotes expressing Flag-Mad2 were cultured for 20–24 h. Metaphase-arrested zygotes were injected with TEV protease mRNA and cultured for at least 2 h. Chromosome spreads were prepared and stained with DAPI to visualize DNA (red) and CREST to mark centromeres (green).
Chromosome spreads prepared from Scc1TEVMyc(m)/+(p) zygotes that had been injected with Flag-Mad2 followed by TEV mRNA confirmed the presence of chromatids (>68 single chromatids and no sister chromatids; n = 6) (Fig. 5B). Interestingly, arm cohesion remained largely intact in Mad2-arrested wild-type zygotes (40 sister chromatids; n = 5), which differs from the loss of arm cohesion in nocodazole-arrested cells (Nakajima et al. 2007). Similar experiments conducted with Rec8TEV(m)/+(p) zygotes revealed that mitotic chromosomes were unaffected by TEV expression (Supplemental Fig. 6B). Consistent with this, unlike Scc1-Myc, Rec8-Myc could not be detected in zygotes (Supplemental Fig. 6C). We conclude that the sister chromatids of zygotes are held together exclusively by cohesin complexes containing Scc1, and that there is little or no Scc1 expression from the paternal allele during the first cell cycle. Furthermore, the loss of cohesion in zygotes following TEV injection demonstrates that the lack of effect in oocytes is not due to Scc1TEVMyc/TEVMyc being uncleavable in vivo, proving that Scc1 has little or no role in holding either bivalents or dyads together. We conclude that fertilization in mammals is accompanied by a rapid and complete switch in the cohesion apparatus, from Rec8- to Scc1-containing cohesin complexes.
No cohesin turnover during the growing phase of oocytes
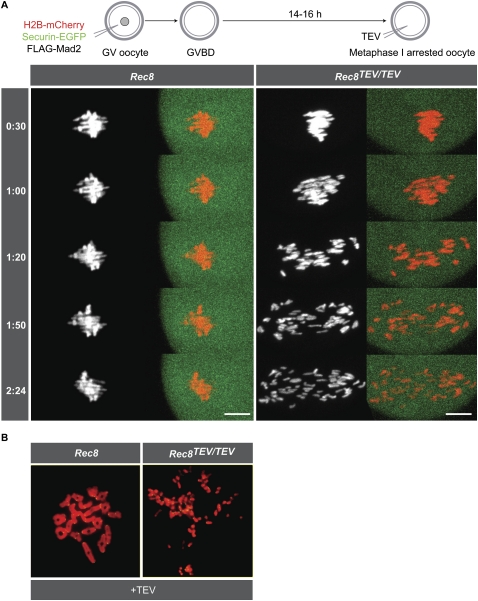
The creation of Rec8TEV/TEV mice enabled us to develop a biological assay for cohesin turnover within the cohesive structures holding bivalents together in oocytes. If non-TEV-cleavable wild-type Rec8 were expressed after the initial establishment of cohesion at DNA replication and there is turnover of cohesin complexes, then the wild-type Rec8 should generate cohesion and bivalent chromosomes should become resistant to TEV (Fig. 6A). To test first whether Rec8 protein translated from mRNA is loaded onto chromosomes, wild-type GV oocytes were injected with Rec8-Myc mRNA carrying 5′ and 3′ globin untranslated regions (UTRs), which are known to promote translation of H2B-mCherry mRNA in oocytes (McGuinness et al. 2009), and cultured in the presence of IBMX for 24 h before fixation. Rec8-Myc was detected using c-Myc-antibody by in situ staining of oocytes in injected but not in uninjected control cells (Fig. 6B). The overexpressed Rec8-Myc was present throughout the oocyte, whereas Scc1-Myc expressed (in different oocytes) from endogenous genes localized predominantly to the GV. Chromosome spreads prepared from oocytes that had expressed Rec8-Myc for 24 h in IBMX and were released into inhibitor-free medium for 5 h revealed little or no chromosomal association, compared with oocytes expressing Rec8-Myc from a BAC transgenic strain, (Tg)Rec8-Myc (Fig. 6C; Kudo et al. 2009).
Because endogenous Rec8 and Scc1 are notoriously difficult to detect on metaphase chromosomes from oocytes and HeLa cells, respectively, the failure to detect Rec8-Myc on chromosomes does not exclude the possibility of new cohesion having been formed. To test if oocytes can generate cohesion under these conditions, Rec8TEV/TEV GV oocytes were injected with H2B-mCherry with or without Rec8-Myc mRNA and cultured for 24 h in IBMX. They were then reinjected with TEV and Securin-EGFP mRNA and released from inhibitor, and chromosomes were observed by live-cell imaging. TEV triggered the conversion of bivalents to chromatids in both control and Rec8-Myc-injected oocytes by 5 h post-GVBD (Fig. 6D), implying that Rec8-Myc cannot form TEV-resistant cohesion under these circumstances.
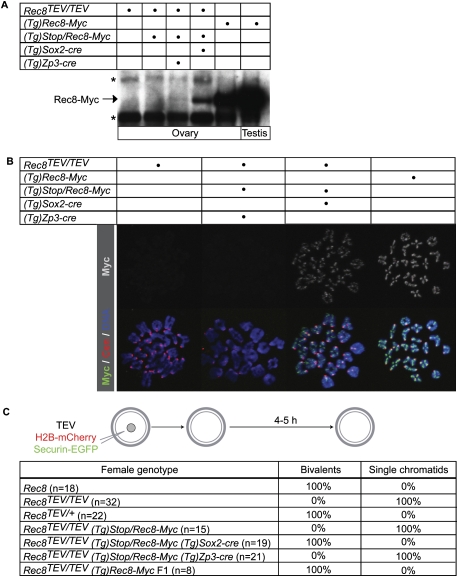
We next used BAC transgenes to control Rec8-Myc expression. By inserting a transcription terminator cassette flanked by LoxP sites (conditional Stop cassette) in intron 1 of Rec8 (Supplemental Fig. 7), expression of Rec8-Myc from such a BAC transgene [(Tg)Stop/Rec8-Myc] can be made conditional on Cre expression (Kudo et al. 2006). By this means, different Cre transgenes can be used to activate Rec8-Myc expression at specific stages of oocyte development. To verify that (Tg)Stop/Rec8-Myc is functional, the Stop cassette was deleted using Sox2-cre, which is expressed in the female germline long before the onset of meiosis. One copy each of (Tg)Stop/Rec8-Myc and Sox-cre rescued not only the infertility of Rec8−/− males and females (litters of seven and nine) but also their postnatal growth defects (Xu et al. 2005) probably caused by inactivation of a gene neighboring Rec8 that is also expressed from (Tg)Stop/Rec8-Myc. Western blotting of extracts from whole ovaries showed that, in the presence of Sox2-cre, (Tg)Stop/Rec8-Myc produced Rec8-Myc protein (Fig. 7A), albeit at lower levels than a multicopy (Tg)Rec8-Myc transgene, which expresses two or more copies of Rec8-Myc constitutively but in a normally regulated manner from the Rec8 promoter (Kudo et al. 2009). Chromosome spreads showed that the protein associated with bivalent chromosomes in oocytes (Fig. 7B). Crucially, it completely suppressed their conversion to chromatids upon injection of Rec8TEV/TEV oocytes with TEV (Fig. 7C). A parallel series of experiments with Rec8TEV/+ oocytes confirmed that bivalent chromosomes are resistant to TEV when only half their Rec8 protein lacks its recognition site (Fig. 7C).
Figure 7.
No cohesin turnover in growing-phase oocytes. (A) Western blot analysis of Rec8-Myc using c-Myc antibody on whole ovary and testis extracts. Rec8-Myc is expressed constitutively from either a multicopy (Tg)Rec8-Myc BAC transgene (second row) or a (Tg)Stop/Rec8-Myc BAC transgene carrying a conditional Stop cassette flanked by LoxP sites (third row) that is activated by Cre recombinase under the control of the Sox2 (fourth row) or Zp3 (fifth row) promoter. The asterisks indicate cross-reacting bands with c-Myc antibody. (B) Chromosome spreads were prepared from four types of oocytes at 5 h post-GVBD. Chromosome spreads were stained with c-Myc antibody to visualize Rec8-Myc (green), CREST to mark centromeres (red), and Hoechst to visualize DNA (blue). (C) Seven types of GV oocytes expressing TEV protease, H2B-mCherry, and Securin-EGFP were cultured in the presence of IBMX for 1 h, then released into IBMX-free culture medium, and chromosomes were visualized by time-lapse confocal microscopy. Oocytes (n = number of cells) were scored as containing either 20 bivalent chromosomes or at least 72 single chromatids (and no bivalents) by 5 h post-GVBD.
Having demonstrated that germline activation of (Tg)Stop/Rec8-Myc prior to meiotic DNA replication creates TEV-resistant cohesion in Rec8TEV/TEV oocytes, we next addressed whether activation at a later stage has a similar effect. To do this, we used the Zona pellucida 3 (Zp3)-driven Cre transgene whose expression during oocyte growth (Lewandoski et al. 1997) causes deletion in primary follicles at the onset of the growing phase (Lan et al. 2004) at least 2–3 wk prior to the formation of mature oocytes. If cohesive structures turn over during this growing phase and are regenerated using protein synthesized de novo, expression of Rec8-Myc from (Tg)Stop/Rec8-Myc induced by Zp3-cre should suppress destruction of bivalents by TEV cleavage (see Fig. 6A).
To test the efficiency with which Zp3-cre removes the transgene's Stop cassette, we genotyped offspring from three mating pairs of Rec8TEV/TEV (Tg)Stop/Rec8-Myc Zp3-cre females (parental generation P0) and Rec8TEV/TEV males. This revealed that 11 out of 11 Rec8TEV/TEV (Tg)Rec8-Myc (with or without Zp3-cre) males and females in the resulting F1 generation lacked the Stop cassette in ear tissue. Since Zp3-cre is expressed exclusively in growing oocytes (Lewandoski et al. 1997), the Stop cassette must have been deleted with high (possibly 100%) efficiency in P0 oocytes prior to fertilization. Western blotting of ovary extracts detected low levels of Rec8-Myc protein expressed from (Tg)Stop/Rec8-Myc due to Zp3-cre (Fig. 7A). The lower abundance induced by Zp3-cre than by Sox2-cre may be partly if not largely due to the fact that only growing oocytes within the ovary will express Rec8-Myc when induced by Zp3-cre, whereas all meiotic cells will express it when induced by Sox2-cre. Despite being expressed, Rec8-Myc induced by Zp3-cre failed to suppress the conversion of bivalents to chromatids upon injection of oocytes with TEV (Fig. 7C). Furthermore, chromosome spreads prepared from Rec8TEV/TEV (Tg)Stop/Rec8-Myc Zp3-cre oocytes failed to detect Rec8-Myc associated with their bivalents (Fig. 7B). Assuming that (Tg)Stop/Rec8-Myc is transcribed upon removal of its Stop cassette by Zp3-cre at levels similar to endogenous Rec8TEV genes, our experiments imply that the cohesion holding bivalents together in mature oocytes is conferred by Rec8 protein synthesized several weeks beforehand; namely, before activation of Zp3-cre.
Discussion
We describe here a novel approach for rapidly inactivating proteins in mammalian cells. It was known that TEV protease could inactivate protein function in budding yeast and more recently in Drosophila (Pauli et al. 2008; Oliveira et al. 2010), but it was unclear whether TEV protease could be used in mammalian cells. We show that TEV protease causes efficient cleavage of Rec8 and Scc1 proteins engineered to contain TEV recognition sites by homologous recombination. TEV protease expression triggers chiasmata resolution and sister centromere disjunction in Rec8TEV/TEV oocytes and eggs, respectively. In the case of the latter, the sister chromatid disjunction induced by TEV takes place in cells arrested in a physiologically normal metaphase arrest—namely, that induced by CSF—proving once and for all that cohesin alone resists spindle forces at this stage of the cell cycle. Our data prove, furthermore, that sister chromatid disjunction during anaphase involves the same fundamental chemistry during meiosis as it does during mitosis. It is important to point out that these conclusions could not have been reached using conventional genetic deletion or RNAi, techniques that interfere with the synthesis of proteins but not directly with their activity. The TEV protease technology provides an alternative to Cre recombinase-mediated gene knockout as a tool for ablating protein function within hours (e.g., a cell cycle phase), and therefore bypasses the potential hazards of secondary pathology resulting from gradual protein rundown. During revision of this paper, it has been reported that a rapamycin-dependent “split” TEV can induce apoptosis in mammalian tissue culture cells (Gray et al. 2010).
Our TEV protease cleavage assay has enabled us to settle several controversial questions. First, the relative importance of cohesin-mediated and cohesin-independent linkages of mammalian chromatids has been called into question (Díaz-Martínez et al. 2007, 2008; Guacci 2007; Yanagida 2009). Our experiments demonstrate that cohesin alone is responsible. Second, the evidence that cohesin holds sister centromeres together during meiosis II in eukaryotic organisms has hitherto been largely indirect; it is based on the simultaneous release of arm and centromere cohesion in mutants lacking the cohesin protection mechanism (Katis et al. 2004, 2010; Kitajima et al. 2004; Rabitsch et al. 2004; Vaur et al. 2005; Riedel et al. 2006; Llano et al. 2008; Ishiguro et al. 2010). Our demonstration that Rec8 cleavage is sufficient to trigger centromere disjunction in metaphase II-arrested eggs settles a long-standing uncertainty and proves that the cohesive structures along chromosome arms and at centromeres are both mediated by Rec8-containing cohesin complexes. Third, we uncovered a dramatic switch between Rec8- and Scc1-containing cohesin complexes at fertilization. Diverse reports that Scc1 mRNA and chromosome-associated Scc1 are present in oocytes led to the notion that Scc1 might also be involved in bivalent cohesion (Lee et al. 2002; Parra et al. 2004; Xu et al. 2004). Unlike Rec8-Myc, we were not able to detect Scc1-Myc on bivalents by chromosome spreads. However, it is clear that large amounts of Scc1-Myc are present in the GV, and minor amounts could, in principle, be involved in either sister chromatid cohesion or conferring mono-orientation of sister kinetochores. Our finding that TEV-induced cleavage of Scc1 has no adverse effects on either meiotic division, while that of Rec8 destroys all cohesion, implies that Scc1 has little or no role during meiotic chromosome segregation. Given that Scc1 becomes vital during the first zygotic division, its accumulation in the oocyte is presumably required to support the first embryonic cell cycle before the onset of zygotic transcription at the two-cell stage. The switchover from Rec8- to Scc1-containing cohesin complexes at fertilization raises interesting questions as to how the molecular footprint of the meiotic program is erased; for instance, whether there is an equally rapid change from shugoshin 2 (Llano et al. 2008) to shugoshin 1 to establish the centromeric protection mechanism required for mitotic chromosome segregation.
Last but not least, our creation of mouse strains with TEV-cleavable Rec8 has enabled us to develop a powerful assay to address whether cohesive structures are continually regenerated from newly synthesized protein during the long prophase arrest of oocytes. Work in yeast suggested that, during unperturbed cell cycles, cohesin cannot build sister chromatid cohesion de novo or exchange subunits of tripartite ring complexes participating in cohesion after the passage of replication forks (Nasmyth and Haering 2009). Whether the same is true of meiotic cells is not known, an issue that is especially relevant in mammalian oocytes where cohesion establishment at DNA replication and destruction at the first and second meiotic divisions can be separated by months in mice and decades in humans. By regulating in a developmental and time-dependent manner activation of a conditional Rec8-Myc BAC transgene, we showed that expression of Rec8 early during meiosis but not during the oocyte growing phase (post-recombination) prevents destruction of bivalents by TEV protease in oocytes whose endogenous genes encode TEV-cleavable Rec8. These data imply that there is little if any significant turnover of Rec8 using newly synthesized protein during the growing phase of oocytes. Future experiments using different transgenes to express the Cre recombinase that activates the Rec8-Myc BAC transgene should enable us to address more precisely if and when de novo Rec8 synthesis can build cohesion; namely, whether it can do so during recombination or only during DNA replication. The lack of Rec8 turnover by our assay is surprising, given that Rec8 forms a complex with Smc1β, which is supposedly loaded by a DNA replication-independent pathway (Eijpe et al. 2003; Hodges et al. 2005). It is conceivable that such a pathway could be responsible for cohesin turnover in resting or growing oocytes.
Our results have important implications for understanding age-related aneuploidy in humans, which, in the absence of an effective turnover mechanism, might reflect progressive degeneration of cohesin complexes holding bivalents together. Consistent with this, a recent cytological study reported an age-dependent reduction of Rec8 and Smc1β in senescence-accelerated mice (Liu and Keefe 2008), although it remains to be demonstrated how reduced levels of chromosome-associated cohesin correlate with rates of aneuploidy. The degeneration of the cohesin complex could be caused by an inadequate synthesis of replacement cohesin proteins, a failure of a turnover mechanism, or a simple time-dependent protein decay without the ability to renew cohesion. A favored hypothesis has been that protein replacement occurs during oocyte arrest or growth (Hodges et al. 2005; Hunt and Hassold 2008). We carefully investigated this and found no evidence for a turnover mechanism that depends on de novo assembled cohesin complexes, at least during the growing phase. We cannot exclude the formal possibility that cohesin complexes forming cohesive structures are somehow recycled from a soluble pool of stable cohesin complexes that do not depend on newly made proteins during oocyte growth. Whether this is indeed the case is extremely difficult to test, and assumes that there is no newly made Smc1β (and Smc3) available to form complexes with Rec8 produced during the growing phase. We therefore propose that the inability of oocytes to regenerate cohesion may indeed contribute to age-related meiosis I errors.
Materials and methods
Gene targeting and generation of mouse strains
For Rec8e10TEV, three consecutive TEV protease recognition sites with a novel MfeI restriction site were introduced between amino acids S274 and P275 of exon 10 by oligo cloning into pFlexible (van der Weyden et al. 2005). Homology arms were retrieved by recombineering using 129Sv-derived BAC 230H5 (Adams et al. 2005) and cloned into pFlexible. The 5′ homology arm consisted of a 6.4-kb ClaI-flanked product (5′ UTR to exon 10). The 3′ homology arm consisted of a 3.8-kb NotI-flanked product (intron 11 to 3′ UTR). To check for homologous recombination, genomic DNA (gDNA) was analyzed by Southern blotting using an external 5′ probe on MfeI-digested DNA to identify a 15.4-kb wild-type allele and a 9.1-kb targeted allele, and using an external 3′ probe on MfeI-digested DNA to identify a 15.4-kb wild-type allele and a 8.9-kb targeted allele.
For Scc1e11TEVMyc, three consecutive TEV protease recognition sites with a novel BamHI restriction site were inserted between P475 and P476 of exon 11 by oligo cloning into pFlexible. Homology arms were retrieved by recombineering using 129Sv-derived BAC 244M24 (Adams et al. 2005). The 5′ homology arm consisted of a 3-kb ClaI-flanked product (intron 8 to exon 11). The 3′ homology arm consisted of a 4.7-kb SbfI-flanked product (3′ UTR). The homology arms were cloned into pFlexible, and a DNA fragment encoding a C-terminal c-Myc10 epitope tag with a Stop codon was inserted by SbfI sites into the targeting vector. To check for homologous recombination, gDNA was analyzed by Southern blotting using a 5′ external probe on BamHI-digested DNA to identify a 13.8-kb wild-type allele, a 7-kb targeted allele lacking the TEV site, and a 4-kb targeted allele including the TEV site, and using a 3′ external probe on BamHI-digested DNA to identify a 13.8-kb wild-type allele, a 9-kb targeted allele lacking the c-Myc tag, and a 6-kb targeted allele including the c-Myc tag.
In vitro culture and microinjection of cells
Fully grown mouse GV oocytes were isolated by physical disaggregation of ovaries of females older than 8 wk but not older than 4 mo in M2 medium (Millipore) supplemented with 200 mM IBMX (Sigma) at 37°C. Oocytes were matured in M16 medium (∼40-μL drops) (Millipore) covered with mineral oil (Sigma) in 5% CO2 at 37°C. Oocytes that had undergone GVBD within 90 min following release from IBMX were selected and cultured further in M16 for experiments.
For zygotes, 3- to 4-wk-old females were injected intraperitoneally with 5 IU of PMSG and 48 h later with 5 IU of hCG, and were paired with B6CBAF1 (Harlan) male mice. Zygotes were retrieved from oviductal ampullae at 17–20 h post-hCG. Cumulus-enclosed zygotes were denuded by exposure to 1 mg/mL hyaluronidase (Sigma) in M2 for 3–6 min, washed through M2, and cultured in M16 in the presence of 5% CO2 at 37°C.
Cells were microinjected in M2 media with a Pneumatic PicoPump (World Precision Instruments) with ∼5–10 pL of mRNA at a final concentration of ∼0.1 μg/mL in RNase-free water (Ambion) into the cytoplasm. GV oocytes were cultured for 1–2 h in M16 supplemented with IBMX to allow for protein expression, and were thereafter cultured in inhibitor-free M16.
Time-lapse live confocal microscopy
Cells were cultured in a PeCon environmental microscope incubator, allowing maintenance of a 5% CO2 atmosphere with humidity at 37°C during time-lapse experiments. A customized Zeiss LSM510 META confocal microscope equipped with P C-Apochromat 63×/1.2 NA water immersion objective lens was used for image acquisition. For detection of EGFP and mCherry, 488-nm and 561-nm excitation wavelengths and BP 505–550 and LP 575 filters were used. Chromosomes labeled with H2B-mCherry were tracked with an EMBL-developed tracking macro (Rabut and Ellenberg 2004) adapted to our microscope. Image stacks of seven to 11 slices of 2–3.4 μm were captured every 2–10 min for 2–7 h.
Acknowledgments
We thank Elizabeth Robertson for Sox2-cre mice, Katja Wassmann for Flag-Mad2 plasmid, and Robin Lovell-Badge and Greg FitzHarris for valuable microinjection advice. We are grateful to Theodore Whipp, Elizabeth Colby, Mayte Siswick, and Magda Wolna for technical assistance. This work was supported by a Medical Research Council grant held by K.N. and K.T.K., and Wellcome Trust and Cancer Research-UK grants to K.N. D.J.A is supported by Cancer Research-UK and the Wellcome Trust, and L.v.d.W is supported by a KKLF Fellowship.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.605910.
Supplemental material is available at http://www.genesdev.org.
References
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, et al. 2005. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758 [DOI] [PubMed] [Google Scholar]
- Bannister LA, Reinhold LG, Munroe RJ, Schimenti JC 2004. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis 40: 184–194 [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387–398 [DOI] [PubMed] [Google Scholar]
- Días-Martínez LA, Giménez-Abián JF, Clarke DJ 2007. Cohesin is dispensable for centromere cohesion in human cells. PLoS ONE 2: e318 doi: 10.1371/journal.pone.0000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Días-Martínez LA, Giménez-Abián JF, Clarke DJ 2008. Chromosome cohesion—Rings, knots, orcs and fellowship. J Cell Sci 121: 2107–2114 [DOI] [PubMed] [Google Scholar]
- Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J Cell Biol 160: 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Feingold E, Dey SK 2009. Etiology of Down syndrome: Evidence for consistent association among altered meiotic recombination, nondisjunction, and maternal age across populations. Am J Med Genet 149A: 1415–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez R, Valdeolmillos A, Parra MT, Viera A, Carreiror C, Roncal F, Rufas JS, Barbero JL, Suja JA 2007. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep 8: 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA 2010. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 142: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V 2007. Sister chromatid cohesion: The cohesin cleavage model does not ring true. Genes Cells 12: 693–708 [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J 2004. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell 15: 951–964 [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P 2001. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2: 280–291 [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P 2007. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet 16: R203–R208 doi: 10.1093/hmg/ddm243 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr Patterns 2: 93–97 [DOI] [PubMed] [Google Scholar]
- Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA 2005. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet 37: 1351–1355 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ 2008. Human female meiosis: What makes a good egg go bad? Trends Genet 24: 86–93 [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y 2010. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 12: 500–506 [DOI] [PubMed] [Google Scholar]
- Johnson MH 1981. The molecular and cellular basis of preimplantation mouse development. Biol Rev Camb Philos Soc 56: 463–498 [DOI] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K 2004. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol 14: 560–572 [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W 2010. Rec8 phosphorylation by casein kinase 1 and Cdc7–Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y 2003. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J 22: 5643–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517 [DOI] [PubMed] [Google Scholar]
- Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, et al. 2006. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 126: 135–146 [DOI] [PubMed] [Google Scholar]
- Kudo NR, Anger M, Peters AH, Stemmann O, Theussl HC, Helmhart W, Kudo H, Heyting C, Nasmyth K 2009. Role of cleavage by separase of the Rec8 kleisin subunit of cohesin during mammalian meiosis I. J Cell Sci 122: 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan ZJ, Xu X, Cooney AJ 2004. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3Cre, and Msx2Cre transgenic mice. Biol Reprod 71: 1469–1474 [DOI] [PubMed] [Google Scholar]
- Lee J, Yokota T, Yamashita M 2002. Analyses of mRNA expression patterns of cohesin subunits Rad21 and Rec8 in mice: Germ cell-specific expression of rec8 mRNA in both male and female mice. Zoolog Sci 19: 539–544 [DOI] [PubMed] [Google Scholar]
- Lee J, Iwai T, Yokota T, Yamashita M 2003. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 116: 2781–2790 [DOI] [PubMed] [Google Scholar]
- Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y 2008. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 10: 42–52 [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 7: 148–151 [DOI] [PubMed] [Google Scholar]
- Liu L, Keefe DL 2008. Defective cohesin is associated with age-dependent misaligned chromosomes in oocytes. Reprod Biomed Online 16: 103–112 [DOI] [PubMed] [Google Scholar]
- Llano E, Gómez R, Gutiérrez-Caballero C, Herrán Y, Sánchez-Martín M, Vázquez-Quiñones L, Hernández T, de Alava E, Cuadrado A, Barbero JL, et al. 2008. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev 22: 2400–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP 1974. Letter: The need for a chiasma binder. J Theor Biol 48: 485–487 [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabé AM, Helmhart W, Kudo NR, Wuensche A, Taylor S, Hoog C, Novak B, et al. 2009. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol 19: 369–380 [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL 1994. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28: 167–187 [DOI] [PubMed] [Google Scholar]
- Murray AW, Szostak JW 1985. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol 1: 289–315 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T 2007. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci 120: 4188–4196 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nicklas RB 1997. How cells get the right chromosomes. Science 275: 632–637 [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat Cell Biol 12: 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TR, Feingold E, Yu K, Cheung V, Tinker S, Yadav-Shah M, Masse N, Sherman SL 2008. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet 4: e1000033 doi: 10.1371/journal/pgen.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MT, Viera A, Gomez R, Page J, Benavente R, Santos JL, Rufas JS, Suja JA 2004. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci 117: 1221–1234 [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson B, Nasmyth K 2008. Cell-type specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 14: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K 2003. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Prieto I, Tease C, Pezzi N, Buesa JM, Ortega S, Kremer L, Martínez A, Martínez-A C, Hultén MA, Barbero JL 2004. Cohesin component dynamics during meiotic prophase I in mammalian oocytes. Chromosome Res 12: 197–213 [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K 2004. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol 14: 287–301 [DOI] [PubMed] [Google Scholar]
- Rabut G, Ellenberg J 2004. Automatic real-time three-dimensional cell tracking by fluorescence microscopy. J Microsc 216: 131–137 [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Yuki K, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan H, Cetin B, et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61 [DOI] [PubMed] [Google Scholar]
- Schultz RM 2002. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod 8: 323–331 [DOI] [PubMed] [Google Scholar]
- Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K 1998. Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ, Harris LW, Tannahill D, Arends MJ, Bradley A 2005. Null and conditional semaphorin 3B alleles using a flexible puroΔtk loxP/FRT vector. Genesis 41: 171–178 [DOI] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM 2003. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol 13: 208–218 [DOI] [PubMed] [Google Scholar]
- Vaur S, Cubizolles F, Plane G, Genier S, Rabitsch PK, Gregan J, Nasmyth K, Vanoosthuyse V, Hardwick KG, Javerzat JP 2005. Control of shugoshin function during fission-yeast meiosis. Curr Biol 15: 2263–2270 [DOI] [PubMed] [Google Scholar]
- Wassmann K, Niault T, Maro B 2003. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol 13: 1596–1608 [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, McKay MJ 2004. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep 5: 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8: 949–961 [DOI] [PubMed] [Google Scholar]
- Yanagida M 2009. Clearing the way for mitosis: Is cohesin a target? Nat Rev Mol Cell Biol 10: 489–496 [DOI] [PubMed] [Google Scholar]