Figure 2. Structural determinants of hERG channel modulators.
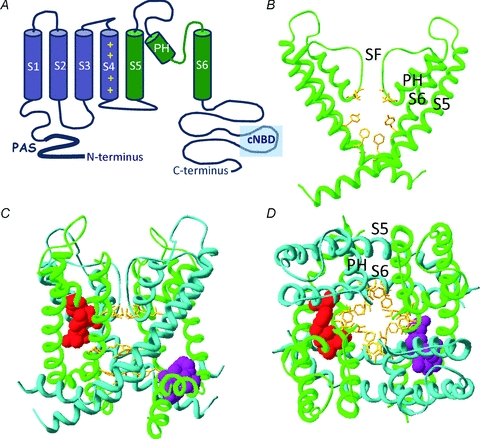
A, cartoon representation of the secondary structure of a single hERG channel subunit. The six transmembrane spanning α-helices (S1–S6) are shown along with the intracellularly located N- and C-termini. The first four helices (S1–S4 shown in blue) form a voltage sensing domain (VSD) which detects changes to the membrane potential. A short linker (S4–S5 linker) connects the VSD to the pore domain, which is composed of the S5 through S6 segments (in green). Within the pore domain are a short α-helix (pore helix, PH) and the selectivity filter (SF), four of which (one from each subunit) come together to coordinate movement of K+ ions through the central pore of the channel. B, homology model of hERG pore domain based upon crystal structure of Kv1.2. Only two of the four subunits are shown for clarity. High-affinity hERG channel blockers bind to amino acid residues (yellow sticks) that line the inner cavity. Two aromatic residues on the S6 helix (Phe656, Tyr652) are critical for all high-affinity blockers. By contrast, the two polar residues on the pore helix (Thr623 and Ser624) are important determinants for some, but not all, high-affinity drugs. C and D, homology model highlighting the putative binding sites for type 1 and type 2 hERG channel activators viewed from the side (C) and extracellular end (D) of the channel. The pore domains from all four subunits are shown by alternating green or blue ribbons. The purple side chains interact with the type 1 activator RPR260243 and are located at the intracellular ends of the S5 helix (Leu553, Phe557) and S6 helix (Asp658, Val659) of a single subunit. In contrast, side chains for the type 2 hERG activator PD118057 are coloured red and are located towards the extracellular end of the S6 helix of one subunit (Leu646) and on the pore helix of a neighbouring subunit (Phe619, Leu622).
