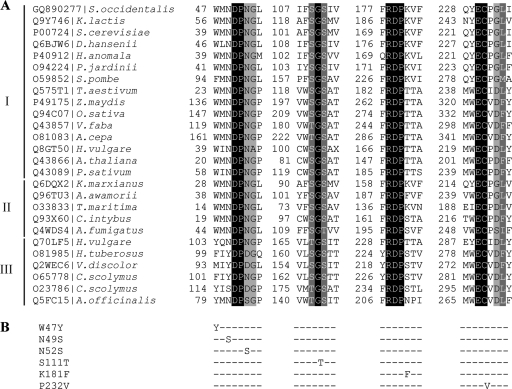
FIG. 4.
(A) Multiple alignments of Ffase and several GH32 proteins in the regions surrounding the sites mutated. The accession code and organism source are indicated in the first column. Identical residues are shaded in black, whereas conserved and semiconserved residues are shaded in dark and light grey, respectively. I, β-fructofuranosidase, EC 3.2.1.26; II, β-fructosidase, EC 3.2.1.80; III fructosyltransferases 2.4.1 (99/100/243); K. lactis, Kluyveromyces lactis; D. hansenii, Debaryomyces hansenii; P. jardinii, Pichia jardinii; S. pombe, Schizosaccharomyces pombe; T. aestivum, Triticum aestivum; Z. maydis, Zea maydis; O. sativa, Oryza sativa; V. faba, Vicia faba; A. cepa, Allium cepa; H. vulgare, Hordeum vulgare; P. sativum, Pisum sativum; K. marxianus, Kluyveromyces marxianus; A. fumigatus, Aspergillus fumigatus; H. tuberosus, Helianthus tuberosus; V. discolor, Viguiera discolor; C. scolymus, Cichorium scolymus; A. officinalis, Asparagus officinalis. (B) Amino acid changes included in the Leu or Ser196 background.