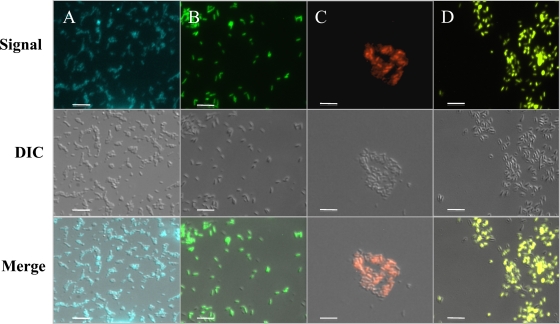
FIG. 2.
Fluorescence microscopy of B. pseudomallei labeled at the att-Tn7 site with gat-cfp (A), gat-gfp (B), gat-rfp (C), and gat-yfp (D). Fluorescent signals were obtained using the respective filter cube sets on a Zeiss AxioObserver D1 microscope and AxioCam MRc 5 monochrome camera. Pseudocolor was applied to the signal intensity at the time of capture using Zeiss AxioVision software. The middle row is comprised of differential interference contrast (DIC) images from the respective samples. At the time of capture, Zeiss AxioVision software was used to superimpose the fluorescent signal and DIC images, displayed in the bottom row. When comparing the overlay in the bottom row to the DIC image in the middle, it can be seen that almost all bacteria are fluorescing at one exposure time or at a set fluorescent intensity. Differing levels of expression of fluorescent protein genes and extended fixation can result in slightly different fluorescent intensities among a population of bacteria, since extended paraformaldehyde fixation could damage the fluorescent proteins (12). The total magnification is ×630, and scale bars equal 10 μm.