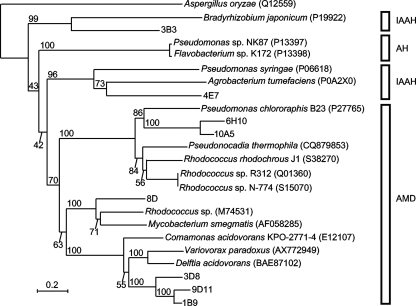
FIG. 3.
Phylogenetic (neighbor-joining) tree of the metagenomic and representative amidases. The topology of the tree was determined by 1,000 bootstrap replicates, and the distances were corrected by the Kimura method (9). Amidases from isolated bacteria are represented with their organismal origin, and accession numbers are in parentheses. The numbers above the nodes represent the bootstrap values. The scale bar indicates the expected number of substitutions per residue. The acetoamidase of Aspergillus oryzae (EA2 subfamily) was used as the outgroup (3). Abbreviations for the distinct enzyme subfamilies are as follows: IAAH, indole-acetamide hydrolases; AH, 6-aminohexanoate-cyclic-dimer hydrolases; AMD, bacterial amidases.