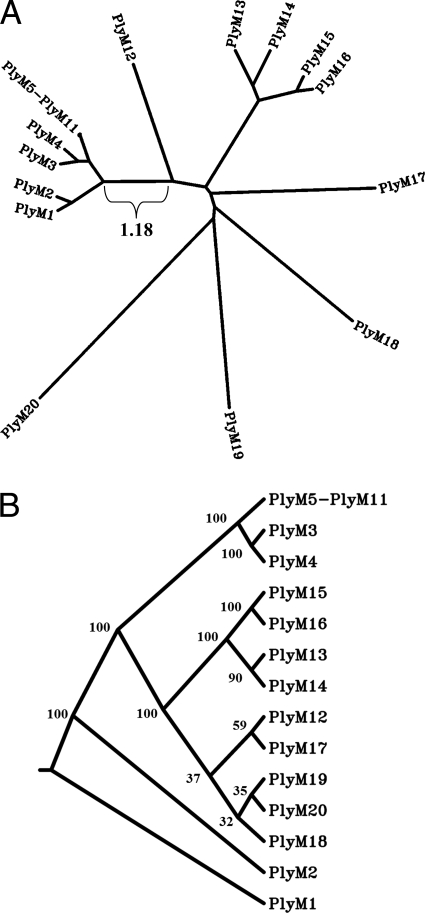
FIG. 3.
Similarities among Gram-positive hits. (A) The sequence homology among the typical Gram-positive lysins (PlyM1 to PlyM20) is organized here as a contemporaneous-tip distance tree (one branch length is included as a reference). Several clusterings are apparent, especially PlyM5 to PlyM11; these enzymes are depicted as a single node in the image, since their degree of homology (90+ pairwise nucleotide/amino acid identity) prevents individual visual differentiation. The reader should note that this tree was generated from a single multiple sequence alignment (MSA) so that the distance values between proteins would be most visually apparent as the branch lengths. (B) Bootstrap analysis (100 rounds) was also conducted on the original MSA of PlyM1 to PlyM20, and a consensus cladogram was generated. As in panel A, PlyM5 to PlyM11 have been collapsed here to a single node. The bootstrap consensus values are indicated at their respective nodes.