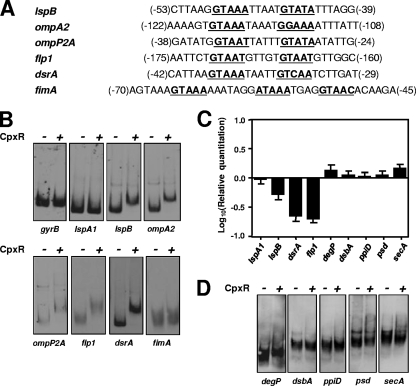
FIG. 4.
CpxR interacts with the promoter regions of genes encoding known and putative H. ducreyi virulence factors but not with the promoter regions of homologs of E. coli cell envelope stress response genes. (A) Nucleotide sequences of the promoter regions for the H. ducreyi lspB, ompA2, ompP2A, flp1, dsrA, and fimA genes. Putative consensus CpxR binding sequences are shown in bold and underlined. Numbers in parentheses indicate the position of the first base of the first repeat (left) and the last base of the last repeat (right) relative to the predicted translation initiation codon. (B and D) Results from EMSAs using 50 pmol of purified rCpxR-His together with the DIG-labeled promoter region DNA. Nucleotide sequences of the primers used to PCR amplify the promoter regions are listed in Table 2. (B) Results from EMSAs using the promoter regions of gyrB, lspA1, lspB, ompA2, ompP2A, flp1, dsrA, and fimA. The lspB, lspA1, and gyrB promoter regions were used as positive (i.e., lspB) or negative (i.e., lspA1 and gyrB) controls in every assay. The minus sign indicates that no CpxR protein was present, whereas the plus sign indicates the presence of CpxR protein. (C) Real-time RT-PCR analysis of gene expression from lspA1, lspB, dsrA, flp1, degP, dsbA, ppiD, psd, and secA in H. ducreyi ΔcpxA relative to wild-type H. ducreyi. (D) Results from EMSAs using the promoter regions of the following H. ducreyi genes: degP, dsbA, ppiD, psd, and secA.