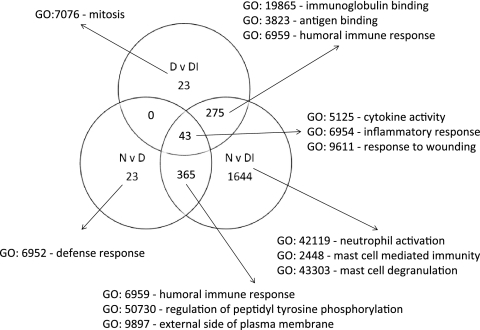
FIG. 2.
Identification of genes with high-degree changes in expression unique to each clinical and infection category, based on >2-fold changes. The biology of each of the gene lists was investigated by testing for enrichment or membership of pathways using DAVID v6, and the major GO terms are listed. The main features reflect those identified based only on high-degree differential expression (Table 2). Influx and activation of cells important in innate immune responses, such as neutrophils and mast cells, are prominent during disease and infection. Humoral and cytokine/chemokine responses typical of adaptive responses appear as expected and are shared between groups. Genes unique to each category were used to generate a small list of genes that could be used to classify participants based on expression profiles and to identify either biomarkers or important expression pathways.