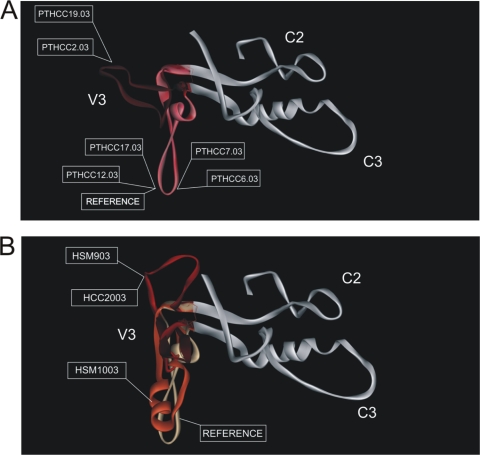
FIG. 4.
Conformational structure of the C2-V3-C3 domains from the primary HIV-2 isolates, as determined by homology modeling. (A) V3 loop conformation of reference HIV-2ALI and neutralization sensitive isolates (PTHCC2.03, PTHCC6.03, PTHCC7.03, PTHCC12.03, PTHCC17.03, and PTHCC19.03). Two different patterns are predicted: a conformational pattern similar to the reference HIV-2 ALI (PTHCC6.03, PTHCC7.03, PTHCC12.03, and PTHCC17.03) and a pattern slightly more exposed than the reference (PTHCC2.03 and PTHCC19.03). (B) V3 loop structure conformation of reference and of neutralization resistant isolates (PTHCC20.03, PTHSM9.03, and PTHSM10.03). All of the conformational patterns are structurally different from the reference HIV-2ALI. The V3 loop is highlighted by a red color gradient, while the C2 and C3 domains are shown in white.