Abstract
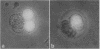
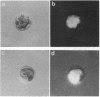
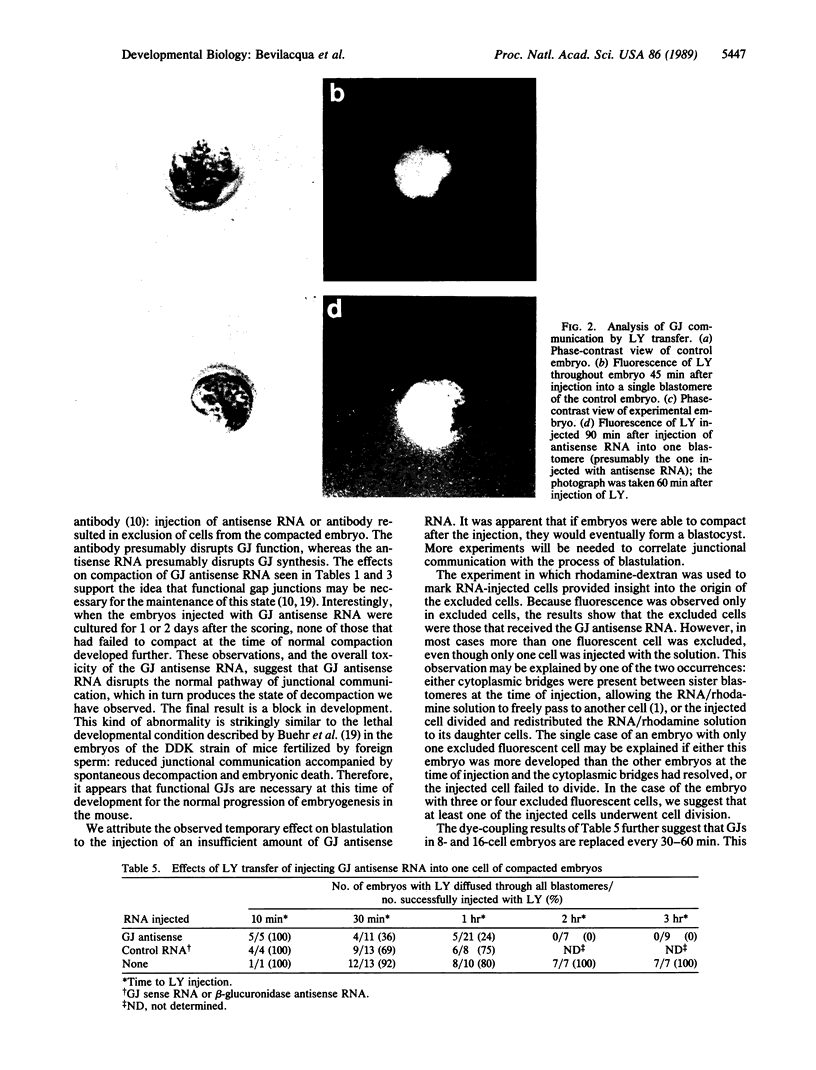
Antisense RNA to the 27/32-kDa rat liver gap junction (GJ) protein was used to explore the role of GJs in preimplantation embryos. When all blastomeres of two- and four-cell embryos were injected with GJ antisense RNA, the percentage of embryos compacted at 60 hr of development was reduced to less than 20%, while 90% of uninjected embryos and 75% of embryos injected with an unrelated RNA were compacted. When most cells of compacted eight-cell embryos were injected with the GJ antisense RNA, 20% of the embryos were decompacted and only 5% had developed to the blastocyst stage at 90 hr, when blastulation had occurred in 90% of the control embryos. When antisense RNA was injected in one blastomere of four-cell embryos, 40% of the embryos presented a large cell that was not included in the compacted embryo at the time of compaction, and an additional 30% of the embryos had two smaller, excluded blastomeres. These excluded cells were identified as the injected cell with a rhodamine-conjugated dextran marker. To assess effects on junctional communication, one blastomere of some embryos was injected with Lucifer yellow, a GJ-penetrating dye, at various times after a blastomere was injected with antisense RNA. The dye was visible in the whole cell mass of control embryos, but it was excluded from a portion of experimental embryos when the delay between the RNA and the Lucifer yellow injections was 1 hr or longer.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ao A., Monk M., Lovell-Badge R., Melton D. W. Expression of injected HPRT minigene DNA in mouse embryos and its inhibition by antisense DNA. Development. 1988 Nov;104(3):465–471. doi: 10.1242/dev.104.3.465. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A., Erickson R. P., Hieber V. Antisense RNA inhibits endogenous gene expression in mouse preimplantation embryos: lack of double-stranded RNA "melting" activity. Proc Natl Acad Sci U S A. 1988 Feb;85(3):831–835. doi: 10.1073/pnas.85.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Lee S., McLaren A., Warner A. Reduced gap junctional communication is associated with the lethal condition characteristic of DDK mouse eggs fertilized by foreign sperm. Development. 1987 Nov;101(3):449–459. doi: 10.1242/dev.101.3.449. [DOI] [PubMed] [Google Scholar]
- Ducibella T., Albertini D. F., Anderson E., Biggers J. D. The preimplantation mammalian embryo: characterization of intercellular junctions and their appearance during development. Dev Biol. 1975 Aug;45(2):231–250. doi: 10.1016/0012-1606(75)90063-9. [DOI] [PubMed] [Google Scholar]
- Fallon R. F., Goodenough D. A. Five-hour half-life of mouse liver gap-junction protein. J Cell Biol. 1981 Aug;90(2):521–526. doi: 10.1083/jcb.90.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall H., Johnson M. H. The nature of intercellular coupling within the preimplantation mouse embryo. J Embryol Exp Morphol. 1984 Feb;79:53–76. [PubMed] [Google Scholar]
- Goodall H., Johnson M. H. Use of carboxyfluorescein diacetate to study formation of permeable channels between mouse blastomeres. Nature. 1982 Feb 11;295(5849):524–526. doi: 10.1038/295524a0. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Constitutive and conditional suppression of exogenous and endogenous genes by anti-sense RNA. Science. 1985 Jul 26;229(4711):345–352. doi: 10.1126/science.2990048. [DOI] [PubMed] [Google Scholar]
- Kalimi G. H., Lo C. W. Communication compartments in the gastrulating mouse embryo. J Cell Biol. 1988 Jul;107(1):241–255. doi: 10.1083/jcb.107.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder G. M., Rains J., McKeon J. Gap junction assembly in the preimplantation mouse conceptus is independent of microtubules, microfilaments, cell flattening, and cytokinesis. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3718–3722. doi: 10.1073/pnas.84.11.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986 Sep;103(3):767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Gilula N. B., Warner A. E. Gap junctional communication and compaction during preimplantation stages of mouse development. Cell. 1987 Dec 4;51(5):851–860. doi: 10.1016/0092-8674(87)90108-5. [DOI] [PubMed] [Google Scholar]
- Lo C. W., Gilula N. B. Gap junctional communication in the post-implantation mouse embryo. Cell. 1979 Oct;18(2):411–422. doi: 10.1016/0092-8674(79)90060-6. [DOI] [PubMed] [Google Scholar]
- Lo C. W., Gilula N. B. Gap junctional communication in the preimplantation mouse embryo. Cell. 1979 Oct;18(2):399–409. doi: 10.1016/0092-8674(79)90059-x. [DOI] [PubMed] [Google Scholar]
- Magnuson T., Demsey A., Stackpole C. W. Characterization of intercellular junctions in the preimplantation mouse embryo by freeze-fracture and thin-section electron microscopy. Dev Biol. 1977 Dec;61(2):252–261. doi: 10.1016/0012-1606(77)90296-2. [DOI] [PubMed] [Google Scholar]
- McLachlin J. R., Caveney S., Kidder G. M. Control of gap junction formation in early mouse embryos. Dev Biol. 1983 Jul;98(1):155–164. doi: 10.1016/0012-1606(83)90344-5. [DOI] [PubMed] [Google Scholar]
- McLachlin J. R., Kidder G. M. Intercellular junctional coupling in preimplantation mouse embryos: effect of blocking transcription or translation. Dev Biol. 1986 Sep;117(1):146–155. doi: 10.1016/0012-1606(86)90357-x. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B., Dermietzel R., Teplow D., Traub O., Willecke K., Revel J. P. Two homologous protein components of hepatic gap junctions. Nature. 1987 Oct 22;329(6141):732–734. doi: 10.1038/329732a0. [DOI] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel J. P., Nicholson B. J., Yancey S. B. Molecular organization of gap junctions. Fed Proc. 1984 Sep;43(12):2672–2677. [PubMed] [Google Scholar]
- Strickland S., Huarte J., Belin D., Vassalli A., Rickles R. J., Vassalli J. D. Antisense RNA directed against the 3' noncoding region prevents dormant mRNA activation in mouse oocytes. Science. 1988 Aug 5;241(4866):680–684. doi: 10.1126/science.2456615. [DOI] [PubMed] [Google Scholar]
- Traub O., Drüge P. M., Willecke K. Degradation and resynthesis of gap junction protein in plasma membranes of regenerating liver after partial hepatectomy or cholestasis. Proc Natl Acad Sci U S A. 1983 Feb;80(3):755–759. doi: 10.1073/pnas.80.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten W. K., Biggers J. D. Complete development in vitro of the pre-implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil. 1968 Nov;17(2):399–401. doi: 10.1530/jrf.0.0170399. [DOI] [PubMed] [Google Scholar]
- Winkel G. K., Pedersen R. A. Fate of the inner cell mass in mouse embryos as studied by microinjection of lineage tracers. Dev Biol. 1988 May;127(1):143–156. doi: 10.1016/0012-1606(88)90196-0. [DOI] [PubMed] [Google Scholar]
- Yancey S. B., Nicholson B. J., Revel J. P. The dynamic state of liver gap junctions. J Supramol Struct Cell Biochem. 1981;16(3):221–232. doi: 10.1002/jsscb.1981.380160303. [DOI] [PubMed] [Google Scholar]