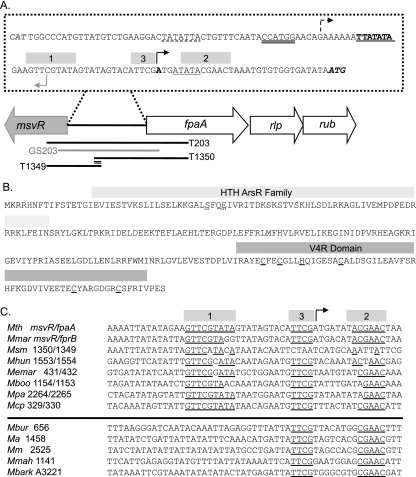
FIG. 1.
(A) Diagram of the M. thermautotrophicus genome region encoding the fpaA-rlp-rub operon (MTH1350 to -1352) and an adjacent transcription regulator (msvR). The sequence of the msvR-fpaA intergenic region is displayed. The PfpaA TATA box is indicated by a solid single underline and bold text, with its corresponding transcription start site identified by a solid black arrow. The PfpaA2 TATA box is represented by a single dashed underline with its putative corresponding transcription start site identified with a dotted arrow. The translation start site for FpaA is shown in italics. The putative PmsvR TATA box is indicated by the thick gray underline, and its putative corresponding transcription start site is identified by a gray arrow. The translation start site for MsvR is shown in gray italics. Potential transcription regulator binding sites are represented by gray boxes. The coverage of templates T203, GS203, T1349, and T1350 is displayed. The overlapping sequence coverage of T1349 and T1350 is double underlined. (B) The amino acid sequence for the putative transcriptional regulator MsvR. Boxes indicate domains identified by the NCBI CDD. The carboxy-terminal amino acid residues that may be important for serving as redox sensors are underlined with a solid black line. N-terminal residues that may be involved in metal binding are represented by a dashed underline. (C) Alignment of the msvR-fpaA intergenic sequence with those of other methanogens (Methanobacteriales and Methanomicrobiales) that have a homologue of the MsvR regulator sharing an intergenic region with a homologue of FpaA. Sequences below the horizontal line are from the promoter regions of msvR genes in members of the Methanosarcinales. The binding regions identified in panel A are displayed here, and conserved nucleotides within these regions are underlined.