Abstract
The cold shock protein (CSP) family includes small polypeptides that are induced upon temperature downshift and stationary phase. The genome of the alphaproteobacterium Caulobacter crescentus encodes four CSPs, with two being induced by cold shock and two at the onset of stationary phase. In order to identify the environmental signals and cell factors that are involved in cspD expression at stationary phase, we have analyzed cspD transcription during growth under several nutrient conditions. The results showed that expression of cspD was affected by the medium composition and was inversely proportional to the growth rate. The maximum levels of expression were decreased in a spoT mutant, indicating that ppGpp may be involved in the signalization for carbon starvation induction of cspD. A Tn5 mutant library was screened for mutants with reduced cspD expression, and 10 clones that showed at least a 50% reduction in expression were identified. Among these, a strain with a transposon insertion into a response regulator of a two-component system showed no induction of cspD at stationary phase. This protein (SpdR) was able to acquire a phosphate group from its cognate histidine kinase, and gel mobility shift assay and DNase I footprinting experiments showed that it binds to an inverted repeat sequence of the cspD regulatory region. A mutated SpdR with a substitution of the conserved aspartyl residue that is the probable phosphorylation site is unable to bind to the cspD regulatory region and to complement the spdR mutant phenotype.
Bacterial stationary phase is characterized by growth arrest as a result of various external causes, such as nutrient starvation, accumulation of toxic compounds, environmental stresses, etc. The mechanisms utilized by the bacterial cell to cope with these phenomena vary accordingly, but in most cases there is a decrease in ribosome activity leading to a severe reduction of protein synthesis. As the cells proceed into longer periods of growth arrest, a reorganization of cell metabolism ensures that the cells will retain viability, maintaining a minimum of essential cell functions (39).
The gene regulation of this passage from exponential growth into stationary phase responds primarily to the nutrient status of the cell. In enterobacteria, the main orchestrator of this shift in metabolism is the sigma factor σS, which substitutes for the vegetative sigma factor σ70 in binding to core RNA polymerase and initiating transcription of a different set of genes (19, 33). The σS-dependent genes include those involved in the stress response as well as metabolic functions, such as uptake and metabolism of amino acids, sugars, and iron and indole production (28). However, no sigma factor with functions equivalent to σS had been identified so far in Alphaproteobacteria.
Another important player is the second messenger guanosine-bis-3′,5′-diphosphate (ppGpp), which is involved in the response to translational stress induced by nutrient starvation (41). In Escherichia coli the levels of this molecule are regulated by two independent enzymes, the ppGpp synthetase RelA and the bifunctional enzyme SpoT, which comprises both ppGpp synthetase and hydrolase activities (20, 51). Other bacterial groups, including the Alphaproteobacteria, contain only one RelA-SpoT homolog, which has both ppGpp synthetase and hydrolase activities (37). It has been demonstrated that ppGpp is important for alternative sigma factor competition for binding the RNA polymerase core during cellular stress (25), causing a reduction in transcription of stable RNAs in favor of amino acid biosynthetic operons and catabolism genes (48).
Among the bacterial proteins that are induced at stationary phase are those containing a cold shock domain (CSD), a protein domain comprising two regions, RNP-1 and RNP-2, that are required for single-strand nucleic acid binding (29, 43). The cold shock protein family includes small polypeptides (7 kDa) that are induced upon temperature downshift and stationary phase (15, 24). Bacterial cells usually have multiple copies of csp genes, and different subsets of these have distinct patterns of expression, being induced by one or more of these stimuli: cold shock, stationary phase, high osmolarity, metal, H2O2, and antibiotics (8, 15, 16, 26, 42, 52). The main proposed role of these proteins is as RNA chaperones maintaining the nucleic acids free from secondary structure at low temperatures, as described for E. coli CspA (24, 26). In addition, some members of this family were proposed to have a role in transcription antitermination of cold-induced genes (E. coli CspA, CspC, and CspE), activation of transcription, translation initiation (Bacillus subtilis CspB), and inhibition of DNA replication (E. coli CspD) (2, 17, 34, 40, 49).
Caulobacter crescentus is a free-living aquatic alphaproteobacterium adapted to low-nutrient conditions, and it has a remarkable cell cycle with an obligatory differentiation step that converts the motile swarmer cell into a sessile stalked cell (23, 46). Its oligotrophic metabolism is ensured by an extensive biosynthetic capability, along with a broad range of pathways for utilization of alternative nutrient sources (9, 21, 38). When the population reaches stationary phase, a large percentage of the cells die, and the ones that survive undergo a morphological change, becoming elongated and having a helicoidal morphology. Cells at this stage are adapted to even more restrictive nutrient conditions and have increased resistance to several stresses (50).
C. crescentus NA1000 has four genes containing cold shock domains; these are cspA and cspB, which are induced by low temperature, and cspC and cspD, which are induced only at stationary phase (31). While the cold-induced CspA and CspB have a single CSD, the two stationary phase-induced genes encode proteins containing two CSDs with molecular masses of 18.3 kDa (CspC) and 21.5 kDa (CspD), a domain arrangement so far found only in Alphaproteobacteria (31). Despite the fact that both proteins have two CSDs, their primary structures are very different, with little amino acid sequence similarity outside the RNP regions, suggesting that they may have overlapping but not identical functions.
Phenotypic analyses of cspC and cspD null mutants, as well as the deletion of both genes, showed that cell viability is severely decreased in the cspC and cspCD mutants upon entry into stationary phase (3). The cspC cells show aberrant morphology at stationary phase, and this phenotype is more severe in the double mutant, suggesting that cspD can at least in part compensate for the lack of cspC (3). This also results from the fact that both cspC and cspD are induced upon entry into stationary phase. The regulatory regions of the two genes have some degree of similarity, and a region upstream of the promoter was shown to be required for maximum levels of expression of cspD (31).
In this work we unveiled the regulatory signals that trigger cspD expression and identified several genes important for its induction at stationary phase; among these signals is a response regulator (SpdR) that belongs to a two-component system with a histidine kinase (SpdS). This is the first report of a two-component system involved in stationary-phase gene regulation in C. crescentus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are described in Table 1. Caulobacter crescentus strains were grown in PYE medium or minimal M2 medium (12) at 30°C with shaking. When necessary, antibiotics were added at the following concentrations: kanamycin, 5 μg/ml; tetracycline, 1 μg/ml; chloramphenicol, 1 μg/ml; or nalidixic acid, 20 μg/ml. Escherichia coli strains were grown at 37°C in Luria-Bertani medium. When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; or tetracycline, 12.5 μg/ml. Plasmids were introduced into C. crescentus by conjugation with E. coli strain S17-1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this work
| Strain, plasmid, or oligonucleotide | Description | Reference or source |
|---|---|---|
| Caulobacter crescentus strains | ||
| NA1000 | Synchronizable derivative of CB15 | 13 |
| SP0200 | NA1000 ΔspoT | This work |
| SP0247 | NA1000 spdR::Tn5 | This work |
| SP0210 | NA1000 ΔspoT ΔspdR | This work |
| ΔspdR mutant | CB15N ΔCC0247::Tetr | 47 |
| ΔspdS mutant | CB15N ΔCC0248::Tetr | 47 |
| E. coli strains | ||
| DH5α | supE44 lacU169 (φ80 lacZΔM15) hsdR17 recA1 endA11 gyrA96 thi-1 relA1 | 18 |
| S17-1 | 294::RP4-2(Tc::Mu)(Km::Tn7) | 45 |
| Plasmids | ||
| pEL4 | pRKlacZ290 with complete cspD regulatory region | 31 |
| pEL5 | pRKlacZ290 with truncated cspD regulatory region | 31 |
| pCA38 | pUJ142 containing the spoT gene | This work |
| pCA39 | pUJ142 containing the spdR gene | This work |
| pCA40 | pUJ142 containing the spdR(D64A) gene | This work |
| pCA401 | pPROEX-HTa containing the spdR(D64A) gene | This work |
| pPROEX-HTa | Expression vector (Ampr) | Invitrogen |
| pGEM-T Easy | Cloning vector (Ampr) | Promega |
| pNPTS138 | Suicide vector containing oriT sacB (Kanr) | D. Alley |
| pSUP2021 | Plasmid containing Tn5 transposon | 45 |
| pUJ142 | Xylose-inducible promoter (Chlorr) | 34 |
| pRKlacZ290 | Transcription fusion vector lacZ, replicon IncP1, oriT (Tetr) | 14 |
| Trx-His CC0247 | Expression vector with cloned coding region of spdR | 47 |
| Trx-His CC0248 | Expression vector with cloned kinase domain of spdS | 47 |
| Cosmid1G4 | pLAFR5 containing nucleotides 1710345-1739376 of the C. crescentus genome | C. Stephens |
| Oligonucleotides | ||
| RELA-1 | GGGGCCCGCGTATCTGAACG | This work |
| RELA-2 | GGGATCCGCTTCGGTCACAGCGGACG | This work |
| RELA-3 | GGGATCCCAAGCACCTGACCAACATC | This work |
| RELA-4 | CCCTGCGGCGCGGAATTCGTCG | This work |
| RELA-5 | GAGGCCTTGTTGGAAGCCGCC | This work |
| CSP2-A | ATTGGATCCATATAACGGCTATGTTCC | This work |
| CAROL-1 | AAGCTTCAAAATCGTAACCAGACATCCC | This work |
| REG-1 | CGAATTCATGGCGGATATCGGAGAACT | This work |
| REG-2 | CTTCGAAGCGAGGGAGCAACTTAAAGC | This work |
| RR-1 | CGGGCCCCCAACTCCAATCTGCTGTGG | This work |
| RR-2 | TGGATCCTCCGCCATAAAAGTCAGCGC | This work |
| RR-3 | CGGATCCAGAAGCGTTTAAGTTGCTCCCTC | This work |
| RR-4 | AGAATTCGTTTGAATTATAATCGGGAGGA | This work |
| MutDA1 | CATGCTGTTCTGGCCATGCGGCTGGAG | This work |
| MutDA2 | CTCCAGCCGCATGGCCAGAACAGCATG | This work |
| R3 | ATGTGACCTCCTAACATGGT | 4 |
| L7 | CCATCTCATCAGAGGGTAGT | 4 |
Complementation and expression vectors.
Plasmid pCA38 was constructed by inserting a 3.8-kb ApaI fragment from cosmid 1G4 containing the spoT gene into vector pUJ142, generating a complementing vector that expresses spoT from its original promoter.
The entire spdR coding region was amplified by PCR using primers REG-1 and REG-2 (Table 1) and cloned into vector pUJ142, generating plasmid pCA39, which contains the spdR gene expressed by the xylX promoter. All the PCRs were carried out with the high-fidelity Pfx enzyme (Invitrogen), using a cloned spdR coding region as the template. A modified spdR coding region was generated by sequential PCR amplification utilizing two complementary mutagenic primers (MutDA1 and MutDA2 [Table 1]) combined with external primers (REG-1 and REG-2) to amplify fragments containing the replacement of the aspartyl codon in position 64 by an alanine codon. The mutant fragment was sequenced to ensure that the correct mutation was introduced and that no additional mutations were generated during amplification. This fragment containing the spdR(D64A) gene was cloned in frame into pUJ142, generating plasmid pCA40, and into the expression vector pPROEX-HTa, generating plasmid pCA401.
Construction of C. crescentus ΔspoT and ΔspoT ΔspdR mutant strains.
A spoT deletion mutant strain was generated by allelic exchange. A fragment of 700 bp located upstream and a fragment of 950 bp located downstream of the spoT gene (32) were amplified by PCR with the primers RELA-1/REL-A2 and RELA-3/RELA-4, respectively (Table 1). The two fragments RELA-1/RELA-2 and RELA-3/RELA-4 were cloned sequentially into the suicide vector pNTS138, and it was introduced into C. crescentus by conjugation with E. coli S17-1. The clones were grown in PYE medium containing 3% sucrose, and those that lost resistance to kanamycin were selected for screening by PCR with primers RELA-4 and RELA-5 (Table 1). One of the clones containing the deletion of spoT obtained after two recombination events was confirmed as the spoT mutant strain SP0200.
A double ΔspoT ΔspdR mutant strain was constructed using SP0200 as the background strain and generating a ΔspdR deletion as described above. Briefly, the flanking regions of spdR were amplified by PCR using primer pairs RR1/RR2 and RR3/RR4 (Table 1), generating amplicons of 950 bp and 970 bp, respectively. The two regions were sequentially cloned into pNPTS138 and introduced in C. crescentus SP0200 to generate a deletion of spdR by double recombination. The resulting ΔspoT ΔspdR strain was named SP0210.
Analysis of cspD expression.
cspD expression was determined by β-galactosidase activity assays of C. crescentus strains NA1000, SP0200, SP0210, and SP0247 harboring plasmid pEL4, which were grown in PYE for 48 h at 30°C with agitation. Strains NA1000 and SP0200 containing plasmid pEL4 were grown in different compositions of M2 medium as follows: M2G (containing 0.2% glucose), M2X (glucose was replaced by 0.3% xylose), M2P (glucose was replaced by 0.2% peptone), or M2GP (containing both glucose and peptone). The cultures were diluted to an optical density at 600 nm (OD600) of 0.1 in the same respective medium, and when the culture was at mid-log phase the expression of the cspD promoter was analyzed at different times of growth by determining β-galactosidase activity as described previously (36). For the assays of glucose and ammonium starvation, cultures of NA1000(pEL4) or SP0200(pEL4) were grown in M2G to mid-log phase. The cells were centrifuged and resuspended in M2 medium containing 0.02% glucose or depleted of NH4Cl. The expression of cspD was analyzed every hour for 5 h and after 24 h by measuring β-galactosidase activity at each time point.
Screening of a transposon mutant library.
A library of 7,500 C. crescentus mutants containing an insertion of the Tn5 transposon (reference 22 and this work) was transferred into PYE plates containing kanamycin and incubated at 30°C for 48 h. E. coli S17-1 containing the plasmid pEL4 was spread over PYE plates and incubated for 2 h at 37°C, and the colonies of the library were transferred on top of the E. coli S17-1 lawn. After growth for 12 h at 30°C, the conjugants were transferred into PYE plates containing kanamycin, tetracycline, and nalidixic acid and incubated at 30°C. The mutants were grown in 150 μl PYE medium in 96-well plates for 2 days, and 4 μl of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (20 mg/ml) was added to the cultures. This experiment used NA1000(pEL4) as a positive control and NA1000(pEL5) (31) as a negative control for the intensity of the blue color developed. The colonies that had a color similar to or lighter than that of the negative control were selected for β-galactosidase assays. Clones in which β-galactosidase activity was less than 50% of that of NA1000(pEL5) were selected for identification of the disrupted gene.
Identification of Tn5 insertion sites.
The Tn5 insertion sites were identified by reverse PCR using primers R3 and L7 as described previously (5). The PCR conditions were as follows: 5 min 95°C; 30 cycles of 1 min at 95°C, 1 min at 42°C or 52°C, and 1 min at 72°C; and a final cycle of 7 min at 72°C. The amplified bands were used for automatic DNA sequencing; the primers used for the sequencing were the same used in the reverse PCR. The identification of the insertion site was determined by comparison to the C. crescentus NA1000 genome at the GenBank database.
Immunoblot analysis.
The polyclonal anti-CspD serum was previously obtained by immunizing New Zealand rabbits (3). Cultures were grown at 30°C in PYE medium for 48 h, and total protein extracts were obtained by sonication followed by centrifugation to remove cell debris. Equal amounts of proteins were separated by electrophoresis in a 15% polyacrylamide-SDS gel and then transferred to nitrocellulose membranes. The membranes were incubated for 16 h with 1:50 anti-CspD antiserum diluted in TBSTT (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.03% Tween 20, 0.02% Triton X-100) and then with anti-rabbit-alkaline phosphatase conjugate (Sigma) at a dilution of 1:5,000 in TBS containing 5% milk. The bands were developed using 0.5 mg/ml nitroblue tetrazolium (NBT) and 0.15 mg/ml 5-bromo-4-chloro-3-indolylphosphate (BCIP) in alkaline phosphatase buffer (100 mM Tris-HCl [pH 9.5], 5 mM MgCl2, 100 mM NaCl).
Expression of the SpdR and SpdS proteins and phosphotransfer assays.
Plasmids Trx-His CC0247 and Trx-His CC0248, with the response regulator SpdR and the histidine kinase domain of SpdS fused with polyhistidine tails (47), were used for overexpression of the corresponding proteins. Plasmid pCA401 was used for overexpression of the mutated SpdR(D64A) protein. The proteins were expressed in E. coli DH5α at 37°C by induction with 300 μM IPTG (isopropyl-β-d-thiogalactopyranoside), and the purification was performed using Ni affinity chromatography (Qiagen).
The phosphotransfer assay was carried out as described previously (47). Briefly, 5 μM purified His-SpdS was autophosphorylated in storage buffer containing 5μCi [γ-32P]ATP at 30°C for 15 min, and then 5 μM purified SpdR was added to the reaction mixture. The phosphotransfer assay was carried out with a reaction mixture containing the histidine kinase alone as a control, and at different times after addition of SpdR (30 s, 2 min, 10 min, and 30 min), the reaction was stopped by the addition of 3.5 μl of 4× sample buffer (500 mM Tris-Cl [pH 6.8], 8% SDS, 40% glycerol, 400 mM β-mercaptoethanol). The samples were separated by 10% SDS PAGE, and the gel was dried and exposed to X-rays film.
Electrophoretic mobility shift assays (EMSA).
The promoter region upstream of the cspD gene was amplified by PCR using primers CSP2-A and CAROL-1 (Table 1). All the following steps were carried out as previously described (10). The probes were end labeled with 20 μCi [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen), and the DNA binding reaction was carried out in 30 μl containing 0.05, 0.1, 0.25, 0.5, and 1 μM purified proteins. Purified His-SpdR or His-SpdR(D64A) proteins were previously incubated with SpdS in the presence of 0.5 mM ATP under conditions described previously (47). In competition assays, a 30-fold excess of unlabeled probe was used to challenge the labeled probe, either of the same fragment (specific competitor) or of a fragment corresponding to the coding region of gene CCNA01712 (nonspecific competitor). After incubation at 30°C for 30 min, the samples were loaded onto a native 5% polyacrylamide gel and electrophoresed in 0.5× Tris-borate (TB) buffer for 2 h at 40 mA. Radioactive species were detected by autoradiography.
DNase I footprinting.
DNase I footprinting assays were performed as described previously (10). The same primers used for amplification of the DNA fragments for EMSA were used for PCR amplification of the cspD promoter region. The reactions and the conditions were the same as used in the EMSA, except that in this case the probe had a single 32P-labeled end. The probe was incubated with increasing amounts of purified His-SpdR protein (0.1, 0.25, and 0.5 μM). The DNA was digested with 0.05 U of RQ1 RNase-free DNase I (Promega) for 1 min. Reaction mixtures were run on a 6% polyacrylamide-urea sequencing gel alongside sequencing ladders of the fragments generated with the same respective primers.
RESULTS
Expression of cspD under different nutrient conditions.
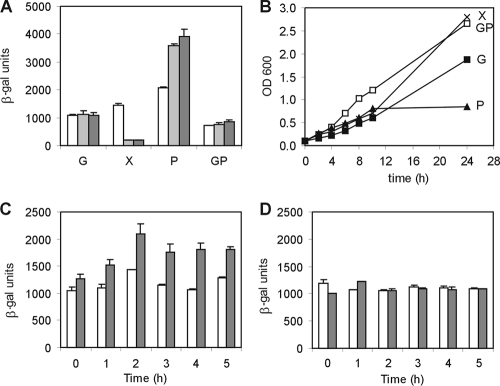
The cspD gene is induced upon entry into stationary phase (31), and previous observations from our group showed that its expression was affected by the medium composition. In order to determine more precisely which nutrient could be affecting expression, we used a transcriptional cspD-lacZ fusion to evaluate cspD expression in media containing different carbon sources. Cultures grown in each medium were freshly diluted in the same medium, and β-galactosidase activity was determined in mid-log phase and early stationary phase (24 h and 48 h). As observed in Fig. 1A, the gene is highly induced in media containing peptone, similar to what was observed in PYE (31), but not in media containing sugars (the gene is not induced in M2G and is repressed in M2X). Moreover, in a medium containing both glucose and peptone, there is a decrease in the maximum levels of expression, suggesting that the presence of glucose could be inhibiting expression. When we analyze the growth of the cultures in each medium, we can observe that growth rates are higher in media containing sugars than in those containing only peptone (Fig. 1B). In fact, an inverse correlation appears to exist between the growth rate and the levels of cspD transcription, and this could be independent of the carbon source utilized.
FIG. 1.
Expression of cspD in C. crescentus NA1000. (A) β-Galactosidase activity assay with wild-type strain NA1000 harboring plasmid pEL4. The cells were grown in different media: G, M2G (containing 0.2% glucose); X, M2X (glucose was replaced by 0.3% xylose); P, M2P (glucose was replaced by 0.2% peptone); GP, M2GP (containing both glucose and peptone). Cultures were incubated at 30°C, and the expression of cspD was determined at mid-exponential phase (white bars), after 24 h (light gray bars), and after 48 h (dark gray bars). (B) Growth curves of C. crescentus NA1000 harboring pEL4 in different media. (C) Expression of cspD in NA1000(pEL4) was determined by β-galactosidase activity assays at mid-exponential phase in M2 medium containing 0.2% glucose (white bars) and at several time points after washing and inoculation in M2 medium containing 0.02% glucose (gray bars). (D) β-Galactosidase activity was determined at mid-exponential phase in M2 medium containing NH4Cl (white bars) and at several time points after washing and inoculation in M2 medium without NH4Cl (gray bars). Error bars indicate standard deviations.
A further evaluation of the role of nutrient availability in cspD expression was carried out to determine whether the gene is induced in response to carbon and nitrogen starvation. The cultures were grown in minimal M2 medium to mid-log phase and then washed and resuspended either in the same medium or in M2 with a low glucose concentration (0.02%) or lacking ammonium chloride. As depicted in Fig. 1C and D, cspD was induced by low glucose but not by ammonia depletion, even though growth was arrested in both cases (not shown). These data indicate that cspD gene regulation responds to the levels of the carbon source, being induced at low concentrations, and that expression varies depending on the carbon source available and the growth rate.
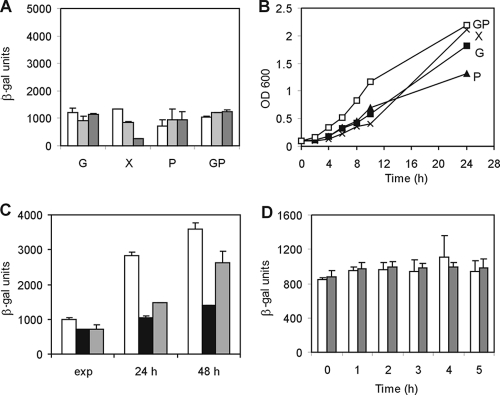
In order to separate the effects of carbon source and growth rate, we then analyzed the expression of cspD in a spoT null mutant strain. In C. crescentus, as in other bacteria, SpoT is the only enzyme responsible for ppGpp synthesis and degradation (32), so we constructed a mutant strain containing a spoT deletion (SP0200) which has the expected phenotype of relaxed growth upon nutrient starvation (compare growth in M2-peptone in Fig. 1B and 2B). A spoT strain carrying a plasmid containing the cspD-lacZ construct was then grown in the same media as the wild-type NA1000 strain (Fig. 1), and expression was measured by β-galactosidase assay. As depicted in Fig. 2A, the induction of cspD at stationary phase in medium containing peptone is no longer observed. This can be confirmed by immunoblot analysis of CspD in the spoT mutant, where very low levels of protein are observed after 48 h of growth in PYE (Fig. 3D). When we analyzed the growth curves in each medium, we can see that the growth rates are high in all media (Fig. 2B), which again correlates inversely to the levels of cspD expression. The induction of cspD in M2-peptone medium was restored when the spoT gene was added back to strain SP0200 (Fig. 2C). Moreover, the induction observed in the shift to low-glucose medium does not occur in the spoT strain (Fig, 2D), indicating that it is dependent on SpoT-mediated signaling.
FIG. 2.
Expression of cspD in the SP0200 strain. (A) β-Galactosidase activity assay was carried out with strain SP0200 harboring pEL4. The cells were grown in different media: G, M2G (containing 0.2% glucose); X, M2X (glucose was replaced by 0.3% xylose); P, M2P (glucose was replaced by 0.2% peptone); GP, M2GP (containing both glucose and peptone). Cultures were incubated at 30°C, and the expression of cspD was determined at mid-exponential phase (white bars), after 24 h (light gray bars), and after 48 h (dark gray bars). (B) Growth curves of SP0200(pEL4) in different media. (C) Complementation of the spoT mutation on cspD expression. Expression of cspD was determined by β-galactosidase activity assays of C. crescentus strains NA1000 (white bars), SP0200 (black bars), and SP0200(pCA38) (gray bars) harboring pEL4. Assays were performed at mid-exponential phase (exp) and stationary phase (24 h and 48 h). (D) Expression of cspD in SP0200(pEL4) was determined by β-galactosidase activity assays at mid-exponential phase in M2 medium containing 0.2% glucose (white bars) and at several time points after washing and inoculation in M2 medium containing 0.02% glucose (gray bars). Error bars indicate standard deviations.
FIG. 3.
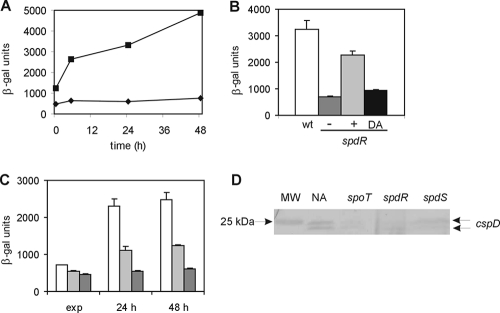
Expression of cspD in spoT and spdR mutant strains. (A) β-Galactosidase activity assay of cspD expression in strains NA1000 (squares) and SP0247 (diamonds) carrying vector pEL4 in PYE medium after different times of growth. (B) Complementation of the spdR mutation on cspD expression. β-Galactosidase activity assay was carried out with C. crescentus strains NA1000 (white bars), SP0247 (dark gray bars), SP0247(pCA39) (carrying the wild-type [wt] spdR gene) (light gray bars), and SP0247(pCA40) [carrying the mutated spdR(D64A) gene] (black bars) harboring pEL4. Assays were performed at stationary phase (48 h). (C) β-Galactosidase activity assay of cspD expression in strains NA1000 (white bars), SP0200 (light gray bars), and SP0210 (dark gray bars) carrying vector pEL4 in PYE medium at exponential phase (exp) and stationary phase (24 h and 48 h). (D) Immunoblot analysis using a polyclonal anti-CspD antiserum of total protein extracts obtained from 48-h cultures of the NA1000, spoT, spdR, and spdS strains grown for 48 h. NA, NA1000 extract; spoT, SP0200 extract; spdR, ΔspdR mutant strain (47) extract; spdS, ΔspdS mutant strain (47) extract.
Taken together, these results suggest that cspD is induced when cells enter a slow-growth period in response to low nutrient levels, particularly carbon sources. The reduced expression of cspD in the spoT strain does not allow us to define whether its transcription is directly activated by the second messenger ppGpp or whether it is responding to another signal transduction network affected in this mutant.
Identification of genes important for cspD expression.
With the purpose of identifying the players in the regulatory systems involved in cspD expression, a library of 7,500 transposon mutants was screened in a search for mutants with reduced cspD expression at stationary phase. The plasmid containing the cspD-lacZ fusion was introduced into the mutant strains by conjugation, and the cultures were grown for 48 h in PYE medium. Expression was evaluated by adding X-Gal to the culture medium, and clones that showed a blue color lighter than that of the positive control were selected for further analysis. Expression of cspD in these selected clones was then quantitatively determined by β-galactosidase activity assays. Ten clones that showed a reduction of at least 50% in the levels of expression were subject to identification of the disrupted gene and are shown in Table 2.
TABLE 2.
Expression of cspD in mutant strains of C. crescentus
| Straina | Disrupted ORFb | Putative ORF productc | β-Galactosidase activity (Miller units)d |
|---|---|---|---|
| NA1000(pEL4) | Wild type | 3,698.7 ± 33.5 | |
| NA1000(pEL5) | Wild type | 740.1 ± 21.0 | |
| 23/11C | CCNA_00247 | Response regulator | 623.0 ± 28.5 |
| 22/7H | CCNA_00254 | RarD family membrane permease | 1,980.3 ± 52.8 |
| 5/3 | CCNA_01611 | Transporter, drug/metabolite exporter family | 2,121.7 ± 125.9 |
| 1/6G | CCNA_02106 | TonB-dependent outer membrane receptor | 1,977.8 ± 120.0 |
| 5/4E | CCNA_02186 | Acetolactate synthase 3 regulatory subunit | 1,740.0 ± 37.7 |
| 24/5E | CCNA_02188 | Cytosolic protein | 1,998.6 ± 47.9 |
| 4/2C | CCNA_02603 | Phosphatidylserine decarboxylase | 1,903.8 ± 5.6 |
| 2/6F | CCNA_02670 | ABC transporter ATP binding protein | 1,022.3 ± 22.5 |
| 46/2A | CCNA_03269 | Glucokinase | 1,863.6 ± 149.2 |
| 30/3C | CCNA_03839 | Acylamino acid-releasing enzyme | 2,102.6 ± 4.8 |
The promoter constructs used are shown in parentheses. All mutant strains contained pEL4.
Numbers indicate the open reading frame (ORF) designation for C. crescentus NA1000 as deposited in GenBank.
Obtained from the sequence databases.
Values are averages and standard deviations of data obtained in three experiments. Data refer to measurements at 24-h time point.
Four of the genes encode proteins involved in transport across the membranes that belong to different systems, i.e., an ABC transporter (CCNA_02670), a permease (CCNA_00254), a TonB-dependent receptor (CCNA_02106), and a putative export system (CCNA_01611). Two independent insertions on neighboring genes (CCNA_02186 and CCNA_02188) also caused a decrease in cspD expression. CCNA_02186 encodes the small subunit of acetolactate synthase, an enzyme involved in the biosynthesis of leucine, isoleucine, and valine, and the product of CCNA_02188 contains a thioesterase superfamily domain. A gene encoding a glucokinase (CCNA_03269) is also important for cspD expression. A strain with a mutation in this gene is still able to grow in M2 containing glucose, but at a lower rate than the wild-type strain, and it shows a similar growth rate in PYE (data not shown). This indicates that this enzyme is probably not the main glucokinase of C. crescentus (a paralog is CCNA_02133), but it confers a better fitness to cells growing with glucose as the sole carbon source. A mutant with a mutation in a gene encoding an acylamino acid-releasing enzyme (CCNA_03839), which is involved in peptide hydrolysis, was also identified in our screen. When analyzing them altogether, we can identify a common feature to these mutations, which is that they disrupted genes that are probably important for nutrient uptake or carbon metabolism. This is in accordance with the fact that cspD is induced by glucose starvation, indicating that an imbalance in the carbon metabolism might interfere with the signals that are important for cspD induction.
Expression of cspD at stationary phase was particularly low in one of the mutants, which was disrupted in a gene encoding a putative response regulator (CCNA_00247). This gene was then named spdR, for stationary-phase cspD regulator, based on evidence obtained in this work (see below). cspD expression was determined at the exponential and early stationary phases (24 h and 48 h) in both wild-type strain NA1000 and the spdR mutant strain SP0247 (Fig. 3A). Expression was lower in the mutant even at exponential phase (time zero), and failed to increase when cells reached stationary phase, keeping a basal level of transcription. The expression of cspD is partially restored in the spdR strain containing the spdR gene in trans under the control of a xylose promoter (Fig. 3B). It should be noticed that in the expression experiments no xylose was added to the medium due to the catabolite repression effect in the presence of xylose (Fig. 1A), so spdR expression was due to the natural leakage of the xylX promoter. In order to evaluate the contributions of SpdR and SpoT to cspD regulation, an spdR spoT double mutant strain was constructed. As can be observed in Fig. 3C, expression of cspD in the double mutant shows a further decrease, suggesting that the two regulators have additive roles in cspD regulation. While cspD expression in the spoT mutant still increases slightly in stationary phase, the spdR mutant (Fig. 3A) and the double mutant (Fig. 3C) do not show this induction, suggesting that SpdR is the major contributor to the stationary-phase induction of cspD. The spdR gene is probably cotranscribed with a gene encoding a membrane-spanning histidine kinase (CCNA_00248), which was here named spdS. Immunoblot analysis of CspD was carried out in spdR and spdS deletion strains at stationary phase (48 h of growth) (Fig. 3D). The results showed that there is very little CspD protein in the spdR and spdS mutant strains, although the decrease in protein amount was less striking in the spdS strain.
Roles of SpdR and SpdS in activation of cspD transcription.
The SpdR protein has a putative conserved aspartyl residue (D64) that could receive a phosphoryl group from a cognate histidine kinase. In order to establish whether SpdS is able to autophosphorylate and to transfer its phosphate to SpdR, a phosphorylation assay was carried out (Fig. 4). Incubation of SpdS with [32P]ATP led to incorporation of the gamma phosphate by SpdS in an autophosphorylation reaction that is typical for this sort of enzyme (47). Addition of purified SpdR to the phosphorylated SpdS caused a very efficient transfer of the labeled phosphate, with detection of labeled SpdR after 0.5 min. These results indicate that SpdS is the cognate histidine kinase of SpdR, and they probably work together regulating genes in response to signals sensed outside the cytoplasmic membrane.
FIG. 4.
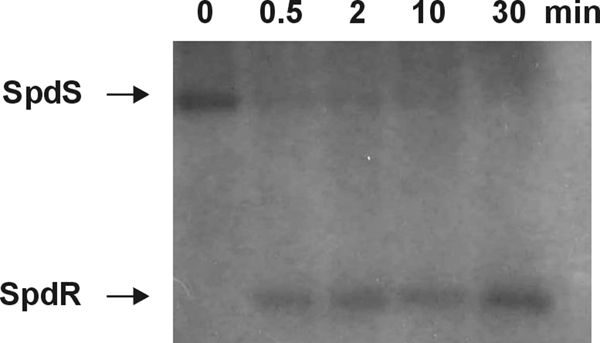
Phosphate transfer between SpdS and SpdR. The purified kinase domain of SpdS was previously autophosphorylated with [32P]ATP and then was mixed with purified His-SpdR. The reaction was stopped at several time points, and phosphotransfer to SpdR was evaluated by SDS-PAGE. Lane 1, autophosphorylated SpdS (time zero); lanes 2 to 5, products of the phosphotransfer reaction after 0.5, 2, 10, and 30 min, respectively.
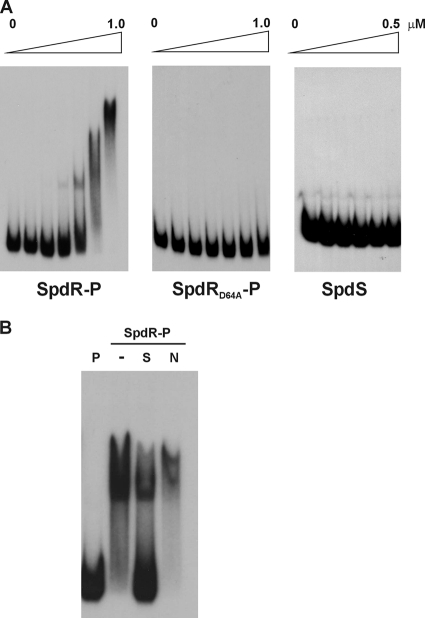
The levels of β-galactosidase activity driven by the complete cspD promoter in the spdR mutant are similar to those obtained with a promoter fusion that lacks the upstream activation region (31) (Table 2), suggesting that SpdR could bind directly to this region and activate transcription of cspD. Binding of purified His-SpdR protein was assessed by electrophoretic gel mobility shift assay (EMSA), using as the probe a fragment containing the complete cspD regulatory region. As shown in Fig. 5A, the probe was shifted in the presence of from 50 nM up to 1 μM His-SpdR that was previously phosphorylated in vitro, and a more pronounced retardation was observed with the increase in protein concentration. This result suggests that the SpdR protein is able to oligomerize when binding to the probe. As a control, purified His-SpdS at the same concentrations was not able to shift the probe. The specificity of SpdR binding was demonstrated in a competition assay, using His-SpdR that was previously phosphorylated in vitro (Fig. 5B). The shifted band was competed out by excess unlabeled DNA probe (specific competitor) but not by an unlabeled nonspecific DNA. Therefore, EMSA experiments confirmed in vitro specific binding of the phosphorylated SpdR regulator to the cspD regulatory region.
FIG. 5.
SpdR binding to the regulatory region of cspD. (A) A DNA fragment corresponding to the regulatory region of cspD was 32P labeled and incubated or not with increasing concentrations (0.025, 0.05, 0.1, 0.25, 0.5, and 1 μM) of purified protein as indicated. His-SpdR and the mutated version His-SpdR(D64A) were previously incubated in phosphotransfer reactions with SpdS. As a negative control, autophosphorylated SpdS was used alone. (B) Competition assays were carried out with 0.5 μM purified His-SpdR that was previously phosphorylated in vitro by SpdS. P, labeled probe; −, probe incubated with His-SpdR; S, probe incubated with His-SpdR in the presence of a 30-fold excess of unlabeled specific probe; N, probe incubated with His-SpdR in the presence of a 30-fold excess of nonspecific DNA competitor.
In order to establish the importance of phosphorylation for SpdR binding, the same experiment was carried out using His-SpdR(D64A), which contains a substitution of the aspartyl residue at position 54 for alanine. This is the probable phosphorylated residue of SpdR, as part of a conserved region in most response regulators. As can be observed in Fig. 5A, in this case the protein was not able to shift the probe even when previously subjected to a phosphotransfer reaction, indicating that phosphorylation at this site is required for SpdR binding. The importance of phosphorylation for SpdR activity in vivo was assessed by complementation studies using an expression plasmid carrying the mutated SpdRD64A version of the SpdR-coding region (Fig. 3B). The results showed that the mutated protein failed to complement the phenotype of cspD activation, confirming that phosphorylation of SpdR in the D64 residue is required in vivo for activation of cspD transcription.
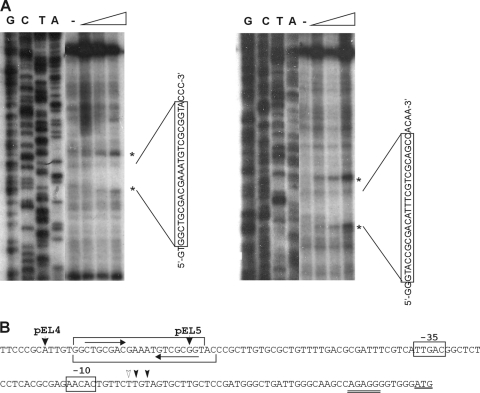
The cspD regulatory region was further analyzed by DNase I footprinting assay to determine the sequence of SpdR binding (Fig. 6). Probes consisting of the cspD promoter region were 32P labeled at the 5′ end of either strand, incubated with increasing amounts of His-SpdR protein, and digested with DNase I. A region corresponding to positions −89 to −66 with respect to the most upstream transcriptional start site of the cspD gene was protected by the SpdR protein in both assays (Fig. 6A). Within this region there is an inverted repeat sequence separated by five nucleotides that could be the motif recognized by SpdR (Fig. 6B). This region had been previously identified as necessary for maximum levels of cspD transcription (31), and our results showed that this is achieved by binding of SpdR and activation of transcription.
FIG. 6.
(A) DNase I footprinting assays of SpdR in the cspD regulatory region. Probes containing the regulatory region of cspD were end labeled and incubated in the presence or absence of increasing concentrations of purified His-SpdR (0.1, 0.25, and 0.5 μM, respectively). The reaction mixtures were treated with DNase I, and reactions were run in a urea-polyacrylamide gel along with a DNA sequencing reaction with the same labeled primer. A minus sign indicates no protein. The protected regions are indicated with the respective sequences, and asterisks mark hypersensitive sites. (B) DNA sequence of the regulatory region of cspD. The −35 and −10 promoter regions are indicated above the sequence. Black arrowheads indicate the transcription start sites, and a white arrowhead indicates the stationary-phase start site of cspD. The start codon is underlined, and the ribosome binding site is double underlined. The SpdR-protected region in each strand is indicated by brackets. An inverted repeat within the SpdR binding site is indicated by arrows. The 5′ ends of the transcriptional fusions pEL4 and pEL5 are indicated.
DISCUSSION
In this work we have studied the regulation of the cspD gene, establishing the signals that trigger its induction at early stationary phase and uncovering part of the regulatory network that modulates its expression. We have observed that the induction of cspD responds to carbon, but not nitrogen, starvation and that it is dependent on the type of carbon source present in the medium. The gene is highly induced in media containing amino acids (peptone) but not in media containing sugars (glucose or xylose), and when cells were grown in xylose the expression of cspD was further repressed at later times of stationary phase. Also, the presence of glucose in the medium containing peptone causes a decrease in the maximum levels of expression compared to those with peptone alone. This effect might be related to catabolite repression, although not much is known about how this is achieved in Caulobacter. Meisenzahl and coworkers reported that repression of lactose utilization of C. crescentus on complex medium can be achieved equally by glucose and xylose, indicating that both sugars may be used as primary carbon sources (35). Moreover, dibutyryl cyclic AMP was shown to stimulate the expression of inducible catabolic enzymes, although the intracellular concentration of cyclic AMP does not vary with carbon source (44).
These results must be taken carefully, since the growth rate of C. crescentus is much higher in media containing sugars than in those containing only amino acids (Fig. 1B). This could indicate that gene induction is somehow responding to the growth rate instead of the carbon source available. This effect was observed earlier in E. coli, where the expression of cspD also varied in response to different carbon sources, and the results indicated that the cspD expression is inversely dependent on the growth rate and independent of the sigma factor σS (52). In fact, circuits modeling gene expression predicted that even the concentration of a protein product of an unregulated gene can exhibit growth rate dependence (27). It is intriguing, however, how the growth rate at exponential phase may interfere with the expression of the gene at the onset of stationary phase. This is more so when we consider that cspD expression is not increased even at later times during stationary phase when cultures have a high growth rate, despite the fact that nutrients should have run out at this point.
When cspD is overproduced in E. coli it causes a lethal effect, and this has been attributed to the fact that it may impair DNA replication, probably by binding to single-stranded DNA (ssDNA) (53). However, increased expression of cspD in C. crescentus does not appear to have the same drastic effect (data not shown), suggesting that it may play a distinct role in the cell physiology and may be regulated differently. Another interesting fact is that C. crescentus CspD appears as two bands in SDS-PAGE, one of them migrating more slowly than predicted by its molecular weight (MW) (3). This indicates that two isoforms of CspD exist in C. crescentus, with one of them likely being subject to posttranslational modification, and this may have an effect upon its function in the cell.
A possible link between the growth rate and cspD expression would be the second messenger ppGpp. In E. coli, ppGpp is produced in response to most stresses and nutrient limitations besides amino acid starvation, and cells grown in minimal medium accumulate more ppGpp than cells grown in rich medium at stationary phase (7, 52). In C. crescentus, deprivation of glucose as the energy source blocks the transition of the stalked cell to the swarmer cell and prevents the beginning of DNA replication. ppGpp was shown not to be necessary for blocking polar morphogenesis, but it is required to inhibit DNA replication, since in the spoT mutant there is an imbalance of the levels of CtrA and DnaA that prevents replication initiation (32).
Expression of cspD in a spoT mutant strain indicated that cspD induction in peptone-containing medium was lost even after 48 h, a time when the nutrients would probably be completely depleted from the medium. Moreover, the carbon starvation induction of cspD was also dependent on the presence of spoT. While in E. coli ppGpp is required for effective transcription of amino acid biosynthetic genes (51), this does not seem to be the case in C. crescentus, since the spoT mutant does not display a phenotype of amino acid auxotrophy (32). Expression of the cspD ortholog in E. coli is very low in the relA spoT double mutant, implying that ppGpp is also a positive factor for the expression of cspD in this bacterium (52). The results indicated that in C. crescentus spoT is required for expression of cspD, either via a direct effect of ppGpp on its transcription or indirectly by the effects on the overall cell response to carbon starvation and growth. In a global analysis of Lactobacillus lactis gene expression in response to carbon limitation (isoleucine or glucose starvation), the stringent response and growth rate were compared, and the results showed that the stringent response is not the general mechanism controlling growth rate modifications or glucose starvation (11).
In this work we have identified several genes that affect the expression of cspD at stationary phase. Most of these genes are involved in nutrient transport and metabolism of carbon compounds, such as amino acids and glucose, in agreement with the fact that cspD is induced upon carbon, but not nitrogen, starvation. Among the genes identified, one encoded a response regulator, SpdR, which was necessary for activation of cspD transcription at stationary phase. Interestingly, the expression of the activator spdR itself was also reported to vary depending on the medium composition (21). An spdR spoT double mutant shows even lower levels of cspD expression, indicating that the effects of each regulator are additive. We could postulate that SpoT is required for signaling for carbon starvation, while SpdR could be involved in signaling for other physiological changes that occur at the onset of stationary phase.
The identification of such a regulator is very important, since very little is known about the control of stationary-phase gene expression in C. crescentus. Cells at this stage are better adapted to cope with very-low-nutrient conditions and have increased resistance to several stresses (50). Some of the genes involved in the increased resistance to stresses at this phase are regulated by extracytoplasmic function (ECF)-type sigma factors, such as some oxidative stress genes that are regulated by the extracytoplasmic sigma factor SigF (1). Moreover, several small regulatory RNAs are induced at stationary phase and by nutrient starvation, suggesting that the regulatory network controlling gene expression at this phase may be more complex than one relying on a single master regulator (30).
SpdR is part of a two-component system and is probably encoded in the same transcriptional unit with the histidine kinase SpdS. Mutants with mutations in these two genes were previously obtained and showed a wild-type phenotype regarding doubling time in PYE, motility, and presence of polar structures (47). We showed that SpdS can phosphorylate SpdR in vitro, in agreement with previous data that histidine kinases display a kinetic preference for their in vivo cognate substrate (47). Moreover, the aspartyl residue D64 was shown to be necessary both for SpdR binding to the cspD promoter (Fig. 5A) and for cspD activation in stationary phase (Fig. 3B). However, CspD protein seems to be present at somewhat higher levels in the spdS mutant strain than in an spdR strain (Fig. 3D), suggesting that other histidine kinases may relay the signal to SpdR. Domain analysis of SpdS showed that it has five transmembrane regions at the amino terminus and a phosphoacceptor and ATP binding domain at the carboxy terminus, indicating that the signal for its phosphotransfer activity could be the binding of an extracellular molecule. Recently attention has been given to auxiliary factors that interact with histidine kinases to modify their activity (6), raising the possibility that SpdS activity could also be the target for a second level of regulation.
The SpdR protein consists of an amino-terminal response regulator domain and a carboxy-terminal helix-turn-helix domain. SpdR binds to an imperfect inverted repeat positioned relatively far upstream from the −35 region of cspD (between positions −87 and −69). This suggests that it may require the formation of dimers or oligomers to allow interaction with the RNA polymerase and activate transcription and that interaction with the carboxyl-terminal domain of the α subunit probably occurs (4). In fact, in vitro DNA binding assays indicate the formation of higher-order structures of SpdR upon interaction with DNA (Fig. 5A). Despite the fact that cspC shows a pattern of expression similar to that of cspD, no SpdR binding sequence can be found upstream of the cspC promoter, and DNA binding assays with purified SpdR did not detect any binding to this region (data not shown), confirming that the regulation of these two csp genes is independently achieved.
The role of SpdR in activating the expression of cspD at the onset of stationary phase suggests that this regulator may be activated by phosphorylation triggered by nutrient starvation, most likely related to the carbon source. The determination of the complete regulon of SpdR and its involvement in stationary-phase adaptation is currently under investigation.
Acknowledgments
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). During the course of this work, C.A.P.T.D.S., H.B., and R.R.M. were supported by fellowships from FAPESP. M.V.M. was partially supported by CNPq.
We thank Michael Laub for the His-SpdR- and His-SpdS-expressing clones and the spdR and spdS mutant strains. We also thank Rogério Lourenço for helping with the His-SpdR and His-SpdS purification and Suely Gomes for critical reading of the manuscript.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Alvarez-Martinez, C. E., R. L. Baldini, and S. L. Gomes. 2006. A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J. Bacteriol. 188:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U. S. A. 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balhesteros, H., R. R. Mazzon, C. A. da Silva, E. A. Lang, and M. V. Marques. 2010. CspC and CspD are essential for Caulobacter crescentus stationary phase survival. Arch. Microbiol. 192:747-758. [DOI] [PubMed] [Google Scholar]
- 4.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 5.Brun, Y. V., and L. Shapiro. 1992. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 6:2395-2408. [DOI] [PubMed] [Google Scholar]
- 6.Buelow, D. R., and T. L. Raivio. 2010. Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75:547-566. [DOI] [PubMed] [Google Scholar]
- 7.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 8.Chanda, P. K., R. Mondal, K. Sau, and S. Sau. 2009. Antibiotics, arsenate and H2O2 induce the promoter of Staphylococcus aureus cspC gene more strongly than cold. J. Basic Microbiol. 49:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, D. K., and A. W. Bourquin. 1987. Metabolism of aromatic compounds by Caulobacter crescentus. J. Bacteriol. 169:1993-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva Neto, J. F., V. S. Braz, V. C. Italiani, and M. V. Marques. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 37:4812-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dressaire, C., E. Redon, H. Milhem, P. Besse, P. Loubiere, and M. Cocaign-Bousquet. 2008. Growth rate regulated genes and their wide involvement in the Lactococcus lactis stress responses. BMC Genomics 9:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 13.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graumann, P., T. M. Wendrich, M. H. Weber, K. Schröder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 16.Graumann, P. L., and M. A. Marahiel. 1999. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis. Arch. Microbiol. 171:135-138. [DOI] [PubMed] [Google Scholar]
- 17.Graumann, P. L., and M. A. Marahiel. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286-290. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogg, T., U. Mechold, H. Malke, M. Cashel, and R. Hilgenfeld. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57-68. (Corrected.) [DOI] [PubMed] [Google Scholar]
- 21.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Italiani, V. C., and M. V. Marques. 2003. Isolation and characterization of Caulobacter mutants impaired in adaptation to stationary phase. Brazilian J. Microbiol. 34:85-90. [Google Scholar]
- 23.Jenal, U. 2000. Signal transduction mechanisms in Caulobacter crescentus development and cell cycle control. FEMS Microbiol. Rev. 24:177-191. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 25.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, P. G., R. A. VanBogelen, and F. C. Neidhardt. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klumpp, S., Z. Zhang, and T. Hwa. 2009. Growth rate-dependent global effects on gene expression in bacteria. Cell 139:1366-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacour, S., and P. Landini. 2004. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landsman, D. 1992. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 20:2861-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landt, S. G., E. Abeliuk, P. T. McGrath, J. A. Lesley, H. H. McAdams, and L. Shapiro. 2008. Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 68:600-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang, E. A. S., and M. V. Marques. 2004. Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain. J. Bacteriol. 186:5603-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesley, J. A., and L. Shapiro. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 190:6867-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mascarenhas, J., M. H. Weber, and P. L. Graumann. 2001. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2:685-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3:585-600. [PubMed] [Google Scholar]
- 38.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyström, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161-181. [DOI] [PubMed] [Google Scholar]
- 40.Phadtare, S., J. Alsina, and M. Inouye. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175-180. [DOI] [PubMed] [Google Scholar]
- 41.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35-51. [DOI] [PubMed] [Google Scholar]
- 42.Schmid, B., J. Klumpp, E. Raimann, M. J. Loessner, R. Stephan, and T. Tasara. 2009. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75:1621-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroder, K., P. Graumann, A. Schnuchel, T. A. Holak, and M. A. Marahiel. 1995. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16:699-708. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro, L., N. Agabian-Keshishian, A. Hirsch, and O. M. Rosen. 1972. Effect of dibutyryladenosine 3′:5′-cyclic monophosphate on growth and differentiation in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 69:1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784-790. [Google Scholar]
- 46.Skerker, J. M., and M. T. Laub. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2:325-337. [DOI] [PubMed] [Google Scholar]
- 47.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traxler, M. F., S. M. Summers, H. T. Nguyen, V. M. Zacharia, G. A. Hightower, J. T. Smith, and T. Conway. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68:1128-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, M. H., A. V. Volkov, I. Fricke, M. A. Marahiel, and P. L. Graumann. 2001. Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription. J. Bacteriol. 183:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wortinger, M. A., E. M. Quardokus, and Y. V. Brun. 1998. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Mol. Microbiol. 29:963-973. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 52.Yamanaka, K., and M. Inouye. 1997. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka, K., W. Zheng, E. Crooke, Y. H. Wang, and M. Inouye. 2001. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39:1572-1584. [DOI] [PubMed] [Google Scholar]