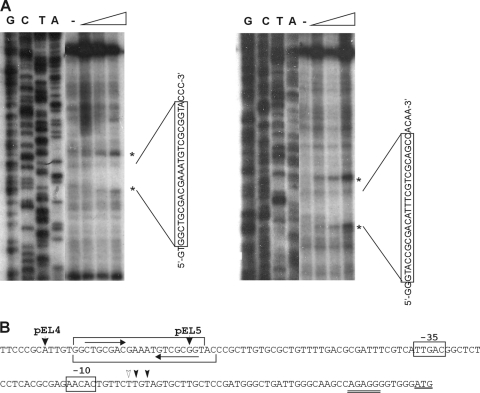
FIG. 6.
(A) DNase I footprinting assays of SpdR in the cspD regulatory region. Probes containing the regulatory region of cspD were end labeled and incubated in the presence or absence of increasing concentrations of purified His-SpdR (0.1, 0.25, and 0.5 μM, respectively). The reaction mixtures were treated with DNase I, and reactions were run in a urea-polyacrylamide gel along with a DNA sequencing reaction with the same labeled primer. A minus sign indicates no protein. The protected regions are indicated with the respective sequences, and asterisks mark hypersensitive sites. (B) DNA sequence of the regulatory region of cspD. The −35 and −10 promoter regions are indicated above the sequence. Black arrowheads indicate the transcription start sites, and a white arrowhead indicates the stationary-phase start site of cspD. The start codon is underlined, and the ribosome binding site is double underlined. The SpdR-protected region in each strand is indicated by brackets. An inverted repeat within the SpdR binding site is indicated by arrows. The 5′ ends of the transcriptional fusions pEL4 and pEL5 are indicated.