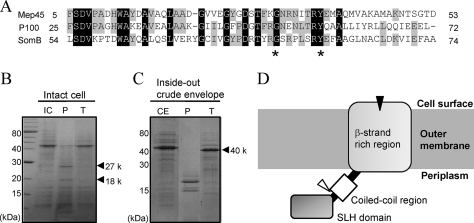
FIG. 1.
Amino acid sequence profiles of Mep45 and determination of the protease-accessible site. (A) Amino acid sequence alignment of the N-terminal SLH domain of Mep45 with the corresponding region of P100 and SomB. Asterisks denote highly and specifically conserved residues found in SLH-bearing outer membrane proteins. (B and C) SDS-PAGE of proteinase K or trypsin treatment of intact cells (B) and crude envelopes (C) of S. ruminantium. Intact cells (3 mg) or crude envelopes (1 mg) were incubated with 100 μg/ml proteinase K or trypsin at 37°C for 30 min. Gels were stained with Coomassie brilliant blue. The bands designated by the arrowheads are the fragments of Mep45 produced by the protease treatment. Abbreviations: IC, intact cells; CE, crude envelopes; P, proteinase K-treated sample; T, trypsin-treated sample. (D) Schematic representation of the topology of Mep45. The locations of proteolytic sites of proteinase K and trypsin are indicated as black and open triangles, respectively. The N-terminal SLH domain protrudes into the periplasmic space, and the C-terminal β-strands span the outer membrane, except for at least one region exposed to the external cell surface, which is the protease-sensitive site.