Abstract
Systemic infections with Elephant Endotheliotropic Herpesviruses (EEHV) cause a rapid onset acute hemorrhagic disease with an 85% mortality rate. More than 60 cases have been confirmed worldwide occurring predominantly in juvenile Asian elephants. Originally, three virus types EEHV1A, EEHV1B and EEHV2 were identified, all members of the Proboscivirus genus within the Betaherpesvirinae. However, four elephant gammaherpesviruses (EGHV) have also been found by DNA PCR approaches in eye and genital secretions of asymptomatic animals, and two more versions of the Probosciviruses, EEHV3 and EEHV4, were recently detected in acute hemorrhagic disease cases. To ask whether even more species of elephant herpesviruses may exist, we have developed several new diagnostic DNA PCR assays using multiple round primers in the DNA POL region. These have been used routinely for nearly three years to screen samples submitted to the Elephant Herpesvirus Laboratory for diagnosis of possible cases of EEHV disease in blood and necropsy tissue, as well as in biopsies of other suspicious lesions or growths. Several more cases of EEHV1-associated hemorrhagic disease were confirmed, but in addition, we describe here eleven examples of other known and novel herpesviruses detected and evaluated with these reagents. They include the prototypes of four new elephant herpesviruses, two more within the Proboscivirus group EEHV5 and EEHV6, plus two more gammaherpesviruses EGHV3B and EGHV5. We also report initial semi-quantitative PCR assays demonstrating very high viral loads in the blood of the EEHV3 and EEHV4-associated hemorrhagic disease cases.
Keywords: Probosciviruses, Gamma Herpesviruses, Hemorrhagic Disease, Mixed Infections, Viral Load
1. Introduction
The first herpesviruses described in association with lethal systemic hemorrhagic disease in elephants were referred to as endotheliotropic herpesviruses (EEHV), because of their morphologically visible effects on infected endothelial cells in necropsy tissue. Phylogenetically, they represent a distinctive third major branch of the Betaherpesvirinae (in addition to the Cytomegalovirus/Muromegalovirus and Roseolovirus genera), and have recently been assigned to a new genus named Proboscivirus (Davison et al., 2009). EEHV is a significant cause of mortality in zoo, circus and sanctuary elephants, affecting predominantly, but not exclusively, juvenile captive-born Asian elephants (Elephas maximus). Overall, more than 60 confirmed cases of EEHV-disease have now been identified, including 33 cases in North America (Richman et al., 1999, Richman et al., 2000a, Garner et al., 2009) and 20 in Europe (Ossent et al., 1990, Ehlers et al., 2001, Fickel et al., 2001, Fickel et al., 2003, Ehlers et al., 2006), as well as several in Asia (Reid et al., 2006) (Zachariah, A, pers comm.). Over 85% percent of these cases have proved fatal. In North America 60% of all deaths of young captive live-born Asian elephants between 4 months to 15 years of age have been attributed to EEHV. The disease has a very rapid clinical course of just one to five days, involving lethargy, edema and tongue cyanosis, as well as microvascular damage, hemorrhaging and focal necrotic lesions in all major internal organs upon pathological examination (Richman et al., 1999, Richman et al., 2000a, Garner et al., 2009). EEHV-like herpesviruses are not known to exist outside of Elephantid hosts and none have yet been isolated by growth in cell culture.
Two related viruses, EEHV1 and EEHV2 were described originally, based on the DNA sequence of two small PCR products U60/Terminase (= TER) and U38/DNA polymerase (POL) after amplification from whole blood and/or necropsy tissue of captive elephants with this disease (Richman et al., 1999). Until recently, EEHV1 has been implicated in all of the cases in Asian elephants, but two known deaths attributed to a similar disease in African elephants (Loxodonta africana) involved EEHV2 instead. Over the past ten years, DNA PCR testing in whole blood samples of live moribund animals or in post-mortem necropsy tissue has been used routinely in our laboratories as a rapid diagnostic assay to successfully detect acute systemic EEHV1 infection (viremia) in 16 more cases of elephant hemorrhagic disease. However, the same PCR test has been uniformly negative in almost all blood samples from healthy animals as well as in numerous unrelated necropsy samples. Based on the results of this PCR test, at least seven systemically infected juvenile Asian elephants that were treated in a timely fashion with the anti-herpesvirus drugs famciclovir or ganciclovir after early suspicious symptoms have survived (Schmitt et al., 2000), although similar treatment was unsuccessful in many other cases. In several of these survivors, the blood PCR test remained positive for three to four weeks, but then declined to below the level of detection thereafter (Richman et al., 2000b).
Genetic analysis of EEHV1 has revealed two major chimeric variants called EEHV1A and EEHV1B, which differ greatly in two non-adjacent genomic loci, including the glycoprotein B (gB) gene region as well as in the glycoprotein-O (gO) plus glycoprotein H (gH) and thymidine kinase (TK) gene block, but are largely very similar otherwise (Ehlers et al., 2001, Fickel et al., 2001, Ehlers et al., 2006) (Richman LK, Zong, J-C, Heaggans, SY, Latimer E and Hayward GS, unpublished data). EEHV1 DNA has also been detected by PCR within both skin nodules and in vulval lymphoid patches from otherwise healthy adult and juvenile African elephants (Jacobson et al., 1986, Munson, 1995, Richman et al., 1999, Richman et al., 2000b). Similarly, EEHV2 DNA was found within the small lung nodules that were reported to be common within healthy culled African elephants (McCully et al., 1971, Richman et al., 2000b). Because of these results, we proposed that the fatal systemic EEHV1-associated disease that predominates in juvenile Asian elephants may involve either direct or indirect cross-species infection with a virus that is endogenous to and generally asymptomatic in African elephants (Richman et al., 1999).
We have recently described the pathologic changes associated with two new cases of fatal hemorrhagic disease in Asian elephant calves that were not attributed to either EEHV1 or EEHV2 (Garner et al., 2009). Instead, we were able to identify two new elephant endotheliotropic herpesvirus genomes in blood and necropsy tissue that we referred to there as EEHV3A and EEHV3B. These are members of a novel G plus C-rich branch (67% GC-content) of the Probosciviruses that is highly diverged from the A plus T-rich branch (42% GC-content) encompassing EEHV1 and EEHV2. However, after additional genetic analysis, those two cases have subsequently proven to be sufficiently distinct to be renamed as EEHV3 and EEHV4 (Richman et al, unpublished data). In both of those cases, two sets of degenerate universal herpesvirus PCR primers, including the TER set used originally to detect EEHV1 (Richman et al., 1999), and the Codehops POL primers used to detect numerous new herpesviruses of veterinary or wildlife interest (VanDevanter et al., 1996, Rose, 2005) each initially generated PCR DNA products from just one of four or five necropsy tissue samples available (colon from #NAP27 and heart from #NAP22). We then also designed a second generation set of more sensitive non-redundant EEHV3-specific TER primers and found that they readily detected the viral DNA by first-round PCR in all five tissue samples from case #NAP27 (EEHV3) and also from all four tissue samples from Case #NAP22 (EEHV4) (Garner et al., 2009).
In addition, a set of four different elephant gammaherpesviruses, which we refer to here as EGHVs, have also been reported in genital and conjunctival swabs of healthy captive elephants in the United States by Wellehan et al (2008). Three of the latter were present in Asian elephants and the fourth in an African elephant. Ehlers et al (2008) have also described an example of one of these same Asian elephant gammaherpesviruses that they termed EmaxGHV1.
Because of the large expansion in the number and genetic diversity of known elephant herpesviruses, we realized the necessity and value of developing several new sets of diagnostic PCR primers designed to improve the sensitivity and specificity for detection of a variety of distinct subsets of elephant herpesviruses. Here we describe a number of additional findings relating to the detection of both known and several previously unknown elephant herpesvirus genomes in either diseased or routine samples collected from captive Asian or African elephants that were forwarded to the National Elephant Herpesvirus Laboratory for analysis. The results presented include the first semi-quantitative measurements of viremia levels in any cases of elephant hemorrhagic disease, the first demonstrations of the presence of EGHVs in localized lesions, nodules or blood samples, as opposed to in either genital or conjunctival swabs, as well as the first reports of multiple herpesvirus infections in a single elephant. In addition, we report the discovery of three more new elephant herpesvirus species, two in the A plus T-rich branch of the Probosciviruses (EEHV5 and EEHV6) and a fifth species of elephant gammaherpesvirus (EGHV5), which is most closely related to whale and dolphin gammaherpesviruses. Another new gammaherpesvirus referred to as EGHV3B is also reported from an African elephant, which is significantly different from EGHV3A found in Asian elephants, but these may not be sufficiently different to justify separate species status. This now brings the total number of known distinct species of elephant herpesviruses to 11 (or 13 if the two related sub-species pairs EEHV1A/EEHV1B and EGHV3A/EGHV3B are included). However, only the Probosciviruses have been associated with acute systemic infection and microvascular damage in cases of fatal hemorrhagic disease, whereas none of the EGHVs have yet been suspected of contributing to systemic disease.
2. Materials and methods
2.1. EEHV and EGHV positive pathological samples
All examples of North American EEHV Probosciviruses have been assigned a chronological NAP number. Those that are relevant to this paper are listed in Table 1. Note that the Proboscivirus isolate numbers used match those of the EEHV disease Case Numbers from which they were obtained. The index case was Kumari (EEHV1 #NAP11) a captive-born female that died at 16 months of age at the National Zoological Park in Washington, DC in 1995. EEHV1A #NAP18 was obtained from a 2.5 year old captive-born male Asian elephant that was born in Missouri and died in California in 2000. The index African elephant case EEHV2 #NAP12 occurred in a captive-born male calf that died at 11 months of age in California in 1996. The chimeric EEHV1B/1A #NAP19 virus was obtained from a 2.5-year old captive-born male Asian calf that died in Missouri in 2002. EEHV1B #NAP14 was found in a 12-year old captive born male Asian elephant born in Texas that died in 1998 in Germany. EEHV3 #NAP27 and agammaherpesvirus sample EGHV2 #NAG1 were both identified in a six-year-old captive-born female Asian elephant that died in 2007 in Washington State. EEHV4 #NAP22 was detected in a five-year old captive-born female Asian elephant in Oklahoma that died in 2004. Gammaherpesviruses EGHV3A #NAG2 and EGHV2 #NAG4 were both detected in DNA extracted from a biopsied vulval lesion from a living 5-year-old captive-born female Asian elephant in 2006 in Florida. Gammaherpesvirus EGHV3B #NAG3 and EGHV2 #NAG5 were both detected in whole blood from a living 22-year-old wild-born female African elephant that had vulval lesions in Tennessee in 2005. The novel gammaherpesvirus EGHV5 #NAG6 was found in a biopsy DNA sample taken from a papillomatous nodule present inside the trunk of a healthy 27-year-old wild-born male Asian elephant in Ohio in 2007. EGHV3A #NAG7 was found transiently at low levels in the blood of a 4-year old male Asian calf with oral discoloration symptoms in Texas in 2009. The novel Proboscivirus genome EEHV5 #NAP28 was detected within two consecutive routine whole blood samples drawn two weeks apart from a living 59-year wild-born female Asian elephant without overt symptoms in Oct 2007. Another novel Proboscivirus genome EEHV6 #NAP35 was detected in a blood sample collected from a 15-month-old African elephant calf in Arkansas that survived after mild illness symptoms and treatment with famciclovir in Mar 2009. Positive EEHV1 control samples used include necropsy tissue DNA from case #NAP21 a three year-old female Asian elephant from New York State in 2003, and from case #NAP20, a 42-year-old, wild-born Asian elephant in Illinois in 2002.
Table 1.
Summary Features of Eleven Elephant Herpesviruses Detected in these Studies.
| Virus Name | Strain/Case Designation | Host Animal Species,Sex,Age | Location | Pathology Description | Tissue Source | DNA Size | Accession Number |
|---|---|---|---|---|---|---|---|
| 1. EEHV3 | #NAP27 | EM, F, 6y | Washington | Fatality | Necropsy tissue (+ blood) | 486-bp | EU658936 |
| 2. EGHV2 | #NAG1 | * | Same | Same | Whole blood | 271-bp | HM060766 |
| 3. EEHV4 | #NAP22 | EM, F, 5y | Oklahoma | Fatality | Necropsy tissue (+ blood | 499-bp | EU658934 |
| 4. EGHV3A | #NAG2 | EM, F, 5y | Florida | Vulval lesions | Whole blood | 171-bp | HM060767 |
| 5. EGHV2 | #NAG4 | * | Same | Same | Same | 271-bp | HM060769 |
| 6. EGHV3B | #NAG3 | LA, F, 22y | Tennessee | Vulval lesions | Biopsy | 457-bp | HM060768 |
| 7. EGHV2 | #NAG5 | * | Same | Same | Same | 271-bp | HM060770 |
| 8. EGHV3A | #NAG7 | EM, M, 4y | Texas | Oral lesions | Whole Blood | 172-bp | HM060772 |
| 9. EGHV5 | #NAG6 | EM, M, 27y | Ohio | Trunk nodule | Biopsy | 457-bp | HM060771 |
| 10 EEHV5 | #NAP28 | EM, F, 69y | Wash, DC | Routine | Whole blood | 463-bp | HM060764 |
| 11 EEHV6 | #NAP35 | LA, F, 15mth | Arkansas | Moribund | Whole blood | 484-bp | HM060765 |
Same case as above
EEHV = Elephant endotheliotropic herpesvirus; EGHV = Elephant gammaherpesvirus; NAP, North American Proboscivirus Case/Isolate #; NAG, North American Elephant Gammaherpesvirus Case/Isolate #; EM = Elephas maximus; LA = Loxodonta africana.
2.2. PCR detection of elephant herpesvirus genomes
The templates for PCR amplification were total DNA samples purified from minced frozen necropsy tissue (Dako Medimachine, Cardinter, CA) or from whole blood using a Gentra Capture Column Kit (Gentra Systems, Minneapolis, MN). The redundant POL PCR amplification employed the following conditions: 34 cycles of 94°C for 1 min, 50 °C for 1 min and 72 °C for 1 min followed by 72 °C for 7 min, using Platinum PCR Supermix (Invitrogen, Carlsbad, CA). All other PCR amplification employed the following conditions: 95°C for 2 min, then 45 cycles of 95°C for 40 sec, 50 °C for 45 sec and 73 °C for 1 min following by 73 °C for 5 min (Promega, Madison, WI). The second generation high efficiency TER primer set specific for EEHV3 (6707/6708/6727/6728) as well as the Pan Herpesvirus Codehops DNA POL primer set (2595/2596/2597) were both described previously (Garner et al., 2009). The descriptions and DNA sequences for six other new sets of EEHV DNA POL PCR primers designed specifically for these studies are listed in Table 2 together with the combinations used and sizes of the products of each primer pair.
Table 2.
Listing of New EEHV DNA POL PCR Primers Used
| Description | Number | Orient | Sequence (5'–3') | Pairing and Product Size (bp) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Round | 1A | 2A | 2B | 3A | |||||
| Specific EEHV3/4: | |||||||||
| 1. EB4P | A1 | 6719 | R | -CGTTGAAGGTGTCGCAGAT - | 390 | 250 | 130 | ||
| 2. EB4P | B1 | 7400 | L | -CAGCATCATCCAGGCCTACAAC - | 390 | ||||
| 3. EB4P | B2 | 6720 | L | -ATCCTGGCGCAGCTGCTGAC - | 250 | ||||
| 4. EB4P | B3 | 6721 | L | -CTCACCTGCAACGCCGTCTA - | 130 | ||||
| PAN EEHV and Specific EEHV6: | |||||||||
| 5. PAN-EBP | A1 | 6710 | R | -ACAAACACGCTGTCRGTRTCYCCRTA- | 500 | 250 | 230 | ||
| 6. PAN-EBP | B1 | 6711 | L | -GTATTTGATTTYGCNAGYYTGTAYCC- | 500 | 470 | |||
| 7. PAN-EBP | B2 | 6712 | L | -TGYAAYGCCGTNTAYGGATTYACCGG- | 250 | 220 | |||
| 8. EB6P | A2 | 7584 | R | - CATCGATTTTGAACTTCTCATGGTC- | 470 | 230 | 220 | ||
| Specific EEHV5: | |||||||||
| 9. EB5P | A1 | 7483 | R | -CTACATCTATACAGAACTTTCC- | 500 | 230 | |||
| 10. EB5P | B1 | 7484 | L | -GTACCTTAGTTACGGACGAGAC- | 500 | 480 | 210 | ||
| 11. EB5P | A2 | 7424 | R | -CGACAGCACACTCCTTTAACAC- | 480 | 210 | |||
| 12. EB5P | B2 | 7485 | L | -CGCTGTATATGGATTTACCGG- | 230 | ||||
| PAN EGHV and Specific EGHV2: | |||||||||
| 13. PANEGP | A1 | 6784 | R | -GTGGKGACTTTGCYAGCCTSTACCC- | 330 | ||||
| 14. PANEGP | B1 | 6788 | L | -ACMCCNGTRAATCCRTASAC- | 330 | 310 | |||
| 15. PANEGP | A2 | 6785 | R | -CCCMAGYATTATWCAGGCMCA- | 310 | 310 | |||
| 16. EG2/3P | A3 | 6786 | R | -CCCCAGCATTATACAGGCACA- | 300 | ||||
| 17. EG2P | B3 | 6787 | L | -ACCCCTGTAAATCCATACACTG- | 300 | 310 | |||
| PAN EGHV and Specific EGHV3: | |||||||||
| 18. PANEGP | A1 | 6784 | R | -GTGGKGACTTTGCYAGCCTSTACCC- | 550 | 330 | |||
| 19. PANEGP | B4 | 7489 | L | -GTCRGTGTCYCCGTAGAYNAC- | 550 | ||||
| 20. PANEGP | A2 | 6785 | R | -CCCMAGYATTATWCAGGCMCA- | 310 | ||||
| 21. EG3P | B5 | 7486 | L | -CTCCCGTAAATCCATACACAGC- | 330 | 310 | |||
2.3. DNA sequencing and phylogenetic analysis
All DNA sequencing was carried out by direct cycle sequencing on both strands of purified PCR DNA products from either one, two or three rounds of nested or semi-nested PCR amplification. The correct sized PCR products were purified after agarose gel electrophoresis with a Qiagen II Gel Extraction kit (Qiagen, Valencia, CA). Sequencing reactions were carried out with the ABI PRISM DigDye Terminator v3.1 cycle sequencing kit and analyzed on an ABI310 DNA sequencer (Applied Biosystems, CA). In other cases, the PCR products were sequenced at the Johns Hopkins School of Medicine DNA Sequencing Core or at Macrogen Inc (Rockville, MD). All DNA sequence editing, analysis and manipulation was performed using Assemblalign and Clustal-W nearest neighbor joining as implemented for MacVector vers 7, together with BLASTX or TBLASTX comparison programs provided online at NCBI.
3. Results
3.1. Further analysis of EEHV hemorrhagic disease cases with EEHV3/4-specific POL primers
Based on the successful large increase in sensitivity obtained with the EEHV3/4-specific TER primers, we set out here to design and test a set of first, second and third-round nested but non-redundant POL gene primers equivalent to the standard 480/520-bp Codehops region that would be expected to be both specific and sensitive for the EEHV4 and EEHV3 versions. Appropriate combinations of these new primers 6719/7400/6720/6721 were positive by first-round PCR with a total of nine tissue samples tested from the fatal Asian elephant calf Cases #NAP27 and #NAP22, as well as from the final whole blood samples of both cases and in an early disease blood sample from #NAP27 (day 4) taken four days before she succumbed (data not shown). As before, the resulting DNA sequences of both of the EEHV3 and EEHV4 POL gene fragments differed from those of EEHV1 and EEHV2 by nearly 30% at the amino acid level, had a G plus C-content of 67% and displayed no more than 55% identity to any other known herpesviral POL proteins in Genbank. Although the POL DNA sequences obtained from all five tissue samples from Case #NAP27 were identical to each other, and the same was true for all four tissue samples from Case #NAP22, the two sets differed at 38 nucleotides and by 16 amino acids (see Table 3). Because of the significant level of DNA and protein divergence between the two cases (at the U73/OBP and U77/HEL gene loci as well as in U38/POL), we have changed the original designation (Garner et al., 2009) of the virus in case #NAP27 from EEHV3A to EEHV3 and that in Case #NAP22 from EEHV3B to EEHV4 (Richman, et al, unpublished data). Importantly, additional semi-quantitative comparisons with these two primer sets on diluted samples revealed that, although they cross-reacted, the EEHV3 TER primer set was up to ten-fold more sensitive for EEHV3 than for EEHV4 and conversely that the EEHV4 POL specific primer set was also about ten-fold more sensitive for EEHV4 than for EEHV3. Both have been tested down to the third-round nested PCR level on unrelated or suspicious necropsy tissue DNA samples from a total of nine additional captive Asian and African elephant necropsies without finding any further positives.
Table 3.
Genetic Comparisons Among the Different EEHV and EGHV POL Genes*
| Amino Acid and Nucleotide Differences (Number / Percentage) | |||||||
|---|---|---|---|---|---|---|---|
| EEHV1A | EEHV1B | EEHV6 | EEHV5 | EEHV2 | EEHV3 | EEHV4 | |
| 1. EEHV1A | - | 2/ 1.2 | 19/11.8 | 30/18.7 | 31/19.4 | 42/26 | 49/30.6 |
| 2. EEHV1B | 15/3.2 | - | 20/12.5 | 31/19.4 | 32/20 | 43/27 | 50/31.2 |
| 3. EEHV6 | 105/22 | 103/21.4 | - | 30/18.7 | 28/17.5 | 45/28 | 51/31.8 |
| 4. EEHV5 | 140/29 | 134/28 | 136/28.3 | - | 19/12 | 46/28.7 | 50/31.2 |
| 5. EEHV2 | 149/31 | 146/30.4 | 143/30 | 100/20.6 | - | 47/29.4 | 53/33 |
| 6. EEHV3 | 192/40 | 187/39 | 199/41.5 | 200/41.7 | 178/37 | - | 16/10 |
| 7. EEHV4 | 190/39.6 | 190/39.6 | 201/42 | 199/41.5 | 169/35.2 | 36/ 7.5 | - |
| Amino Acid and Nucleotide Differences (Number / Percentage) | ||||||
|---|---|---|---|---|---|---|
| EGHV1 | EGHV2 | EGHV3A | EGHV3B | EGHV4 | EGHV5 | |
| 8. EGHV1 | - | 53/33 | 59/37 | 59/37 | 59/37 | 65/40.6 |
| 9. EGHV2 | 167/34.8 | - | 64/40 | 64/40 | 61/38 | 62/38.7 |
| 10.EGHV3A | 183/38 | 194/40.4 | - | 4/2.4 | 23/14.4 | 45/27 |
| 11.EGHV3B | 185/38.5 | 199/41.4 | 25/ 5.2 | - | 20/12.5 | 45/28 |
| 12.EGHV4 | 185/38.5 | 188/39 | 104/21.7 | 100/20.8 | - | 46/29 |
| 13.EGHV5 | 193/40.2 | 189/39.3 | 162/33.7 | 165/34.4 | 170/35.4 | - |
Codehops region only ( = 480-bp, 160 amino acids from positions 605 to 777)
Case #NAP27 did not show sufficient similarities in overt symptoms to the earlier instances of EEHV disease to trigger administration of famciclovir during the 8 days of her illness, and treatment of Case #NAP22 with famciclovir failed to affect the outcome. Therefore, we considered it important to attempt to estimate the viral load present within their blood samples by limiting dilution PCR. In the absence as yet of quantitative real-time PCR assays for EEHV3 or EEHV4, we prepared a series of eight sequential five-fold dilutions for both, in which the total amount of DNA present was maintained at a constant level by the addition of human placental carrier DNA. In each case, the original undiluted PCR reaction contained 2 μl of DNA from a total 100 μl sample prepared from 200 μl of whole blood. These diluted samples were then amplified by nested PCR with our EEHV3-specific TER primer set (data not shown). The final #NAP27 (day 8) whole blood sample proved to give positive results down to 625-fold and 15,625-fold dilution after first and second-round amplification respectively, but was negative at the next dilution in each case. A similar experiment was also carried out with diluted #NAP22 whole blood using the EEHV4-specific POL primer set, which gave positive results down to 125-fold and 3,125-fold after first and second-round PCR (data not shown). The latter result is very similar to that obtained with whole blood samples from several untreated cases of EEHV1 disease, which have been measured by quantitative real-time PCR to have viral load values of close to 107 viral genome copies per ml (Stanton et al., 2010). Both primer sets failed to amplify any EEHV3 or EEHV4 sequences (even after third-round PCR) from both undiluted whole blood DNA samples taken at the same time as the disease in Case #NAP27 from all three herdmates, including one African and two Asian elephants, as well as from a whole blood sample collected one year prior to disease from the same animal that died (data not shown).
3.2. Detection of an additional low level gammaherpesvirus infection in the case of EEHV3-positive hemorrhagic disease
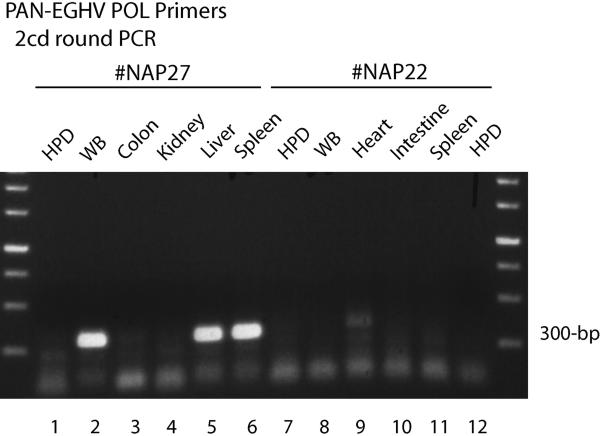
To assess whether any gammaherpesvirues might also be present in elephant necropsy or blood samples, where they could generate complications with regard to diagnosis, we next designed a set of redundant PAN EGHV primers (6784/6785/6788). These were based on comparisons of the available EGHV POL sequences with other mammalian gammaherpesvirus POL DNA sequences, including eliminating mismatches from both the outer and inner Codehops primers. Using a combination of both first-round moderately redundant consensus EGHV primers, followed by a second-round with EGHV2-specific elephant gammaherpesvirus primers (6786/6787), one of these same gammaherpesvirus genomes (designated #NAG1) was also detected within Case #NAP27, the same fatal Asian elephant case that presented with high levels of the Proboscivirus EEHV3. However, positive results for the EGHV genome were only obtained after second-round PCR (using 6785/6787) in undiluted liver, spleen, and intestine samples, and also in the final whole blood sample (Fig 1, lanes 2, 5 & 6), but not in other necropsy tissues tested such as colon and kidney (Fig 1, lanes 3 & 4).
Figure 1. Detection of a gammaherpesvirus POL gene in some tissues of Case #NAP27.
Agarose gel electrophoretic separation of ethidium bromide-stained second-round PCR products obtained using PAN EGHV primers 6784/6788 then EGHV2-specific primers 6786/6787 (300-bp). Lanes1, 8 and 13, human placental DNA negative controls (HPD); Lanes 2 to 7, Case #NAP27 undiluted DNA samples; 2, whole blood; 3, colon; 4, kidney; 5, liver; 6, spleen. Lanes 9 to 12, Case #NAP22 undiluted DNA samples; 9, whole blood; 10, heart; 11, intestine; 12, spleen. The two outside lanes show a 250-bp multimer size marker ladder.
The unique 271-bp POL DNA sequence obtained for this gammaherpesvirus proved to be identical to that previously described for ElHV4 (Wellehan et al., 2008), which we call EGHV2 to discriminate them from the EEHVs and to be consistent within the gammaherpesvirus set (see Table 4). No EGHV PCR products were detected in these samples after 5, 25 or 125-fold dilution. In contrast to Case #NAP27, a similar set of tests on all diluted and undiluted tissue DNA samples from Case #NAP22 were negative after two-round PCR with the same PAN EGHV primer set, except for a weak cross-reacting band from EEHV4 in the heart tissue sample only (Fig 1, lane 9).
Table 4.
List of All Known Species of Elephant Herpesviruses:
| Proposed Name | ICTV Number | Host Species# | Pathology | Reference |
|---|---|---|---|---|
| 1. EEHV1A | ElHV1 | EM | Hemorrhagic disease | Richman, 1999 |
| 2. EEHV1B | * | EM | Hemorrhagic disease | Fickel, 2001 |
| 3. EEHV2 | ElHV2 | LA | Hemorrhagic disease | Richman, 1999 |
| 4. EGHV1 | ElHV3 | EM | Mucosal shedding | Wellehan, 2008 |
| 5. EGHV2 | ElHV4 | EM | Mucosal shedding | Wellehan, 2008 |
| 6. EGHV3A | ElHV5 | EM | Mucosal shedding | Wellehan, 2008 |
| 7. EGHV3B | * | LA | Genital lesion | This study |
| 8. EGHV4 | ElHV6 | LA | Mucosal shedding | Wellehan, 2008 |
| 9. EEHV3 | ElHV7 | EM | Hemorrhagic disease | Garner, 2009 |
| 10.EEHV4 | ElHV8 | EM | Hemorrhagic disease | Garner, 2009 |
| 11.EEHV5 | ElHV9 | EM | Mild viremia | This study |
| 12.EGHV5 | ElHV10 | EM | Trunk nodule | This study |
| 13.EEHV6 | ElHV11 | LA | Mild viremia | This study |
EM = Elephas maximus; LA = Loxodonta africana.
Same as above
3.3. Detection of additional novel elephant herpesviruses with PAN EEHV primers
Because of the large increase in recognized elephant herpesvirus species, we next designed a set of two-round PAN-EEHV POL locus primers 6710/6711/6712, that both encompassed the known expanded sequence information about the EEHV1, EEHV2, EEHV3 and EEHV4 POL loci (Richman et al, unpublished data), and at the same time reduced the level of redundancy and mismatches compared to those in the standard Codehops PAN-herpesvirus primers. The new PAN-EEHV primers still proved capable of detecting both EEHV3 and EEHV4, as well as all other known EEHV positive samples tested by either first or second-round PCR within available positive necropsy tissue samples, including examples of EEHV1A, EEHV1B, EEHV2 and EEHV4 (Fig 2, lanes 1 to 6). However, they were least efficient for EEHV3. This test has now been used over a three-year time period for routine screening of all blood, biopsy and necropsy samples submitted to the NZP Elephant Herpesvirus laboratory. In addition to being effective in detecting several new cases of EEHV1A or EEHV1B-associated hemorrhagic disease (not addressed here), we have also detected eight more examples of other elephant herpesviruses as described below, all within samples from healthy or surviving elephants, but which in most cases had some suspicious illness or observable lesions. Surprisingly, six of these positive samples proved to be elephant gammaherpesviruses, with five being variants of either the EGHV2 or EGHV3 viruses described by Wellehan (Wellehan et al., 2008), whereas the other represented a fifth different and novel species of elephant gammaherpesvirus (EGHV5). In addition, we also identified two more novel Probosciviruses (EEHV5 and EEHV6) with these PAN EEHV primers.
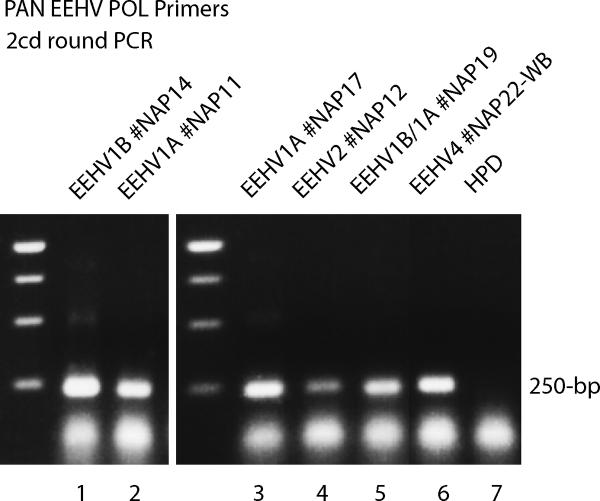
Figure 2. Detection of EEHV1A, EEHV1B, EEHV2 and EEHV4 with PAN EEHV POL primers.
Agarose gel electrophoretic separation of ethidium bromide-stained second-round PCR products obtained using the moderately redundant PAN-EEHV POL Codehops region primers 6710/6711 (500-bp) then 6710/6712 (250-bp). The 250-bp ladders are shown on the left of both the center and right-hand panels. The following necropsy DNA samples were used. Lane 1, EEHV1B case #NAP14; 2, EEHV1A case #NAP11; 3, EEHV1A case #NAP17; 4, EEHV2 case #NAP12; 5, EEHV1B/1A case #NAP19; 6, EEHV4 case #NAP22 whole blood; 7, human placental DNA negative control (HPD). The samples from EEHV1B #NAP11, EEHV1A #NAP17 and EEHV4 case #NAP22 heart were also all strongly positive even after just first-round PCR with primers 6710/6711 (500-bp not shown), whereas those from EEHV1A #NAP11 and EEHV1B/1A #NAP19 were only weakly positive first-round.
3.4. Detection of simultaneous infection with two different gammaherpesviruses in both an Asian and an African elephant with vulval lesions
Positive gammaherpesvirus POL PCR DNA products of 250-bp detected with our PAN-EEHV primers were identified during second-round PCR using 6710/6712 within blood obtained from a healthy 6-year-old captive-born female Asian elephant with vulval lesions (#NAG2), as well as directly in biopsy tissue from a healthy 22-year-old wild-borne African elephant with vulval lesions (#NAG3) (data not shown). Despite the fact that we had intended these primers to preferentially detect all EEHV genomes, analysis of the unique 172-bp DNA sequences obtained here revealed instead two EGHV3 POL genes very similar to that of ElHV5 as reported in swabs from Asian elephants by Wellehan et al (Wellehan et al., 2008). The EGHV3 DNA sequence obtained from our Asian elephant blood sample (#NAG2) proved to be identical to that of the ElHV5 described by Wellehan (2008), but there were 5 nucleotide differences here (2.9%) in the African elephant EGHV3 biopsy sample (#NAG3) compared to that of ElHV5.
Unexpectedly, in both animals with these vulval lesions, we also subsequently detected a second different gammaherpesvirus genome EGHV2 when we attempted to obtain the other segment of the 480-bp Codehops POL PCR locus using our new PAN EGHV primer set. Both of the primer pairs 6788/6786 and 6785/6787 gave 310-bp bands after second-round PCR, in which one primer of each pair preferentially targets EGHV2 (Fig 3, lanes 1 & 4). The Asian elephant EGHV2 (#NAG4) DNA sequence obtained here was again identical to that given for ElHV4 by Wellehan (2008), but the African elephant EGHV2 (#NAG5) sample contained 3 out of 271-bp differences (1.1%) from the Asian version. We were unable to detect the other POL gene segment from either of these two EGHV2 viruses using the double PAN-EGHV primer set 6785/6789 at second-round (Fig 3, lanes 7 & 8), nor were those primers able to detect the known high abundance EEHV1 genome present in a positive necropsy sample from Case #NAP20 (Fig 3, lane 2).
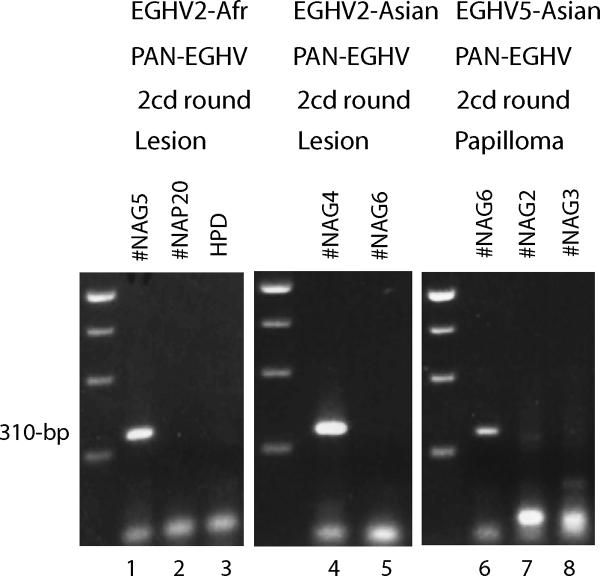
Figure 3. PCR detection of EGHV2 in an African and Asian elephant and EGHV5 in an Asian elephant.
Agarose gel electrophoretic separation of ethidium bromide-stained PCR products obtained using moderately-redundant consensus PAN-EGHV or PAN-EEHV POL primers. Lanes 1 to 5, detection of EGHV2 with first-round PAN-EGHV primers 6784/6788 followed by second-round EGHV2-specific primers 6785/6787 (310-bp). Lanes 1, African elephant #NAG5, vulval lesion; 2, Asian elephant EEHV1-positive case #NAP20; 3, Human placental DNA negative control (HPD); 4, Asian elephant #NAG4, vulval lesion; and 5, Asian elephant #NAG6, trunk papilloma. Lanes 6 to 8, detection of EGHV5 with first-round PAN-EGHV POL primers 6784/6788 followed by second-round PAN-EGHV POL primers 6785/6788 (310-bp); 6, Asian elephant #NAG6, trunk papilloma; 7, Asian elephant #NAG2/4, vulval lesion; and 8, African elephant #NAG3/5, vulval lesion. The three left-hand side lanes contain a 250-bp multimer size marker ladder.
3.5. Evidence for distinctive Asian and African forms of EGHV3
Introduction of another EGHV3-specific primer 7486 used together with the EGHV PAN primer 6785 for second-round PCR after a first-round with 6784/7489 was successful in detecting the other 310-bp segment of the EGHV3 POL gene Codehops region from the African elephant vulval lesion biopsy (#NAG3) (data not shown). The combined 457-bp POL locus DNA sequence displayed 25 nucleotide (5.5%) and 5 amino acid (3.1%) differences from the prototype EGHV3 (ElHV5) POL gene described by Wellehan at al (2008). Therefore, we have tentatively named the apparent African version of this virus as EGHV3B (#NAG3) to distinguish it from the closely related EGHV3A (#NAG2) virus found in both the blood from the Asian elephant with vulval lesions and in the Wellehan studies (see Table 4).
3.6. Transient appearance of EGHV3A in the blood from an Asian elephant calf with oral lesions
Another case occurred in a 4 year-old Asian calf (#NAG7) that was suspected of having early stage EEHV disease and was given pre-emptive famciclovir after displaying oral lesions and tongue discoloration. However, it proved instead to have a low level of EGHV3 present in a single blood sample taken before treatment (but not in samples collected after treatment). This time, a weak 250-bp PCR product band was detected after second-round amplification with the PAN EEHV primers 6710/6712, which proved to be identical to EGHV3A rather than an EEHV (data not shown). We were not able to amplify the other half of the POL codehops region from this sample. Oral lesions and tongue discoloration are often thought to accompany the initial stages of EEHV-associated disease, but this case illustrates that EGHVs need to be considered as well as likely causes of non-hemorrhagic oral lesions.
3.7. Identification of a novel elephant gammaherpesvirus (EGHV5) in a trunk growth
A third different elephant gammaherpesvirus was also detected with our PAN EEHV primer set 6710/6711/6712 after two-round PCR in a biopsy DNA sample obtained from a proliferating papilloma-like growth present inside the trunk of a healthy wild-born 27-year-old male Asian elephant (#NAG6; data not shown). The initial second-round 250-bp DNA sequence obtained from this sample proved to be that of a novel DNA POL gene that clearly fits within the gammaherpesvirus subfamily, but is phylogenetically closer to the POL proteins of bottlenose dolphin gammaherpesvirus and of beaked whale gammaherpesvirus than to any other known herpesviruses. Notably, it is also significantly closer to both of those virus species than to any of the other four known elephant gammaherpesviruses (see Fig 6B). Pathological examination of the papilloma revealed the presence of typical herpesvirus nuclear inclusion bodies and virions within differentiated epithelial cells.
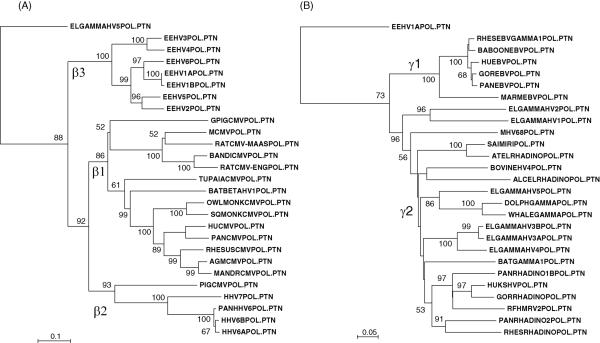
Figure 6. Phylogenetic tree comparisons of the DNA POL genes of elephant herpesviruses with those of other mammalian beta and gammaherpesviruses.
Phylograms of predicted protein coding sequence data for the complete 480-bp and 160 amino acid POL Codehops region (Rose, 2005) from all thirteen species and sub-species of elephant herpesviruses were compared with data for the orthologous regions of selected representative mammalian herpesviruses. These were generated by the neighbor joining best tree procedure after alignment in Clustal-W with Tajima-Nei and Poisson adjustments and show distance based branch lengths as well as bootstrap values (1000 reiterations). (A): Protein level comparisons for 19 other betaherpesviruses with the seven species or subspecies of EEHV using the gammherpesvirus EGHV5 as the outgroup. Genbank accession numbers for the other betaherpesvirus sequences used were as follows: guinea pig (caviid) CMV FJ355434, mouse CMV U68299, rat CMV (Maastricht) AF232689, bandicoot CMV EF125067, rat CMV (English) AY728086, Tupaia (tree shrew) CMV AF281817, bat betaherpesvirus AB517983, owl monkey (Aotes) CMV FJ483870, squirrel monkey (Saimiri) CMV FJ483967, human CMV (Merlin) AY446894, Pan (chimpanzee) CMV (Haberling) AF480884, rhesus macaque CMV AY186194, African green monkey CMV FJ483968, mandrill CMV AF282941, pig (porcine) CMV AF268042, human HHV7 U43400, Pan (chimpanzee) HHV6-like roseolovirus AY359407, human HHV6B AF157706 and human HHV6A X83413. (B): Protein level comparisons for 20 other gammaherpesviruses with the six species or sub-species of EGHV using the betaherpesvirus EEHV1 as the outgroup. Genbank accession numbers for the gammaherpesvirus sequences used were rhesus (macaque) EBV-like lymphocrytovirus AY037858, baboon (Papio) EBV AY037858, human EBV (Epstein-Barr virus B95-8 strain) V01555, gorilla EBV GQ921926, Pan (chimpanzee) EBV AY166457, marmoset EBV AF319782, mouse MHV68 U97553, Saimiri (squirrel monkey) rhadinovirus X64346, Ateles (spider monkey) rhadinovirus AF083424, bovine herpesvirus 4 AF318573, alcelephine rhadinovirus AF005370, bottlenose dolphin gammaherpesvirus DQ288667, beaked whale gammaherpesvirus AY949828, elephant gammaherpesvirus 4 EF531714, bat gammaherpesvirus AB298558, Pan (chimpanzee) rhadinovirus 1B AF250881, human KSHV (HHV8) U75698, gorilla rhadinovirus AF250886, RFHM (macaque) rhadinovirus 2 AF005478, Pan (chimpanzee) rhadinovirus 2 AF290601, rhesus (macaque) rhadinovirus 1 (RV1) AF083501.
This novel EGHV genome represented the ninth elephant herpesvirus species identified and, for the same reasons given above, we prefer to use the more informative name elephant gammaherpesvirus 5 (EGHV5) rather than ElHV9 (Table 4). We were also able to obtain the other remaining 310-bp segment of the EGHV5 POL gene Codehops locus from this same papilloma sample (#NAG6) by using the PAN EGHV primer pair 6785/6788 for second-round PCR after a first-round with 6785/6788 (Fig 3, lane 6). However, this combination was not successful for any of the vulval lesion EGHV2 and EGHV3 genomes (#NAG2, 3, 4 and 5; Fig 3, lanes 7 and 8). EGHV5 was also not detected when the EGHV2-specific second-round primer pair 6786/6787 was used (Fig 3, lane 5). The combined DNA sequences from these two smaller PCR products extended the total known size of the EGHV5 POL locus up to a 457-bp sequence of 42% G plus C-content encoding a protein with 65, 62, 45, and 46 amino acid differences from those of the orthologous segments of the EGHV1, EGHV2, EGHV3 and EGHV4 POL proteins, respectively (Table 3). Therefore, although very highly diverged, EGHV5 is somewhat more closely related to the EGHV3/EGHV4 branch than to the EGHV1/EGHV2 branch, displaying 73% residual amino acid identity to the former but only 60% identity to the latter.
3.8. Identification of EEHV5: a novel Proboscivirus that is most closely related to EEHV2
A fourth example of an elephant herpesvirus (isolate #NAP28) identified with our PAN EEHV primers 6710/6711/6712 was found when carrying out routine PCR blood test screening from a 59-year-old wild-born female Asian elephant, that had shown some signs of mild illness nine months earlier and occasionally displayed small oral lesions. Initially, 250-bp second-round PCR products (data not shown) were obtained with the 6710/6712 primers from two sequential blood samples collected over a two-week period, but not from a third at one month later. Evaluation of the DNA sequence of these two PCR products revealed identical but novel POL genes that were more closely related to the EEHVs than to any other known herpesviruses, but were also distinctly different from all other previously identified EEHV POL genes. Therefore, we have designated this new elephant herpesvirus as EEHV5.
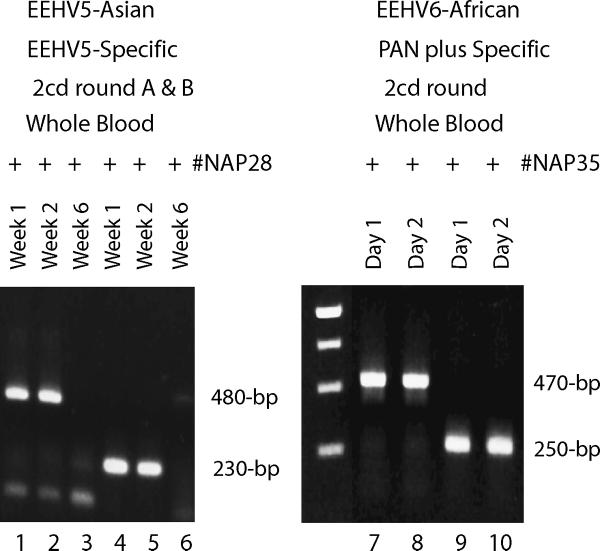
The EEHV5 viral DNA was of low abundance and difficult to detect consistently even after second-round PCR with the PAN EEHV primers. Therefore, we designed second-generation EEHV5-specific PCR primers and successfully expanded the known EEHV5 POL region to the more standard 480-bp Codehop region size, to permit more robust phylogenetic analysis. Two alternative second-round primers pairs from the final chosen primer set of 7483/7484/7424/7485 produced strong PCR bands of 480-bp and 230-bp from both the first and second week blood samples from #NAP28 (Fig 4, lanes 1, 2, 4 and 5): however, again the one month later blood sample from the same animal was negative (lanes 3 and 6). The combination of data from the two PCR products yielded a 499-bp DNA sequence with a G plus C-content of 43% similar to that of EEHV1 and EEHV2, but with 19 (12%) and 31 (19%) amino acid differences from EEHV2 and EEHV1 respectively (Table 3). Subsequent evaluation with these same specific primers of 24 sequential routine blood samples drawn from this elephant over the previous two-years proved to be negative (data not shown) implying that this animal had undergone a transient but low-level asymptomatic systemic reactivation event.
Figure 4. PCR detection of two additional novel endotheliotropic herpesviruses (EEHV5 and EEHV6).
Agarose gel electrophoretic separation of ethidium bromide-stained PCR products obtained using consensus PAN and specific EEHV POL primers. Lanes 1 to 6, detection of EEHV5 in two of three successive whole blood DNA samples from Asian elephant #NAP28 over a six week time course; lanes 1 to 3, EEHV5-specific first-round primers 7483/7484 then EEHV5-specific second-round primers 7424/7484 (480-bp); 4 to 6, first-round EEHV5-specific primers 7483/7484 then alternative second-round EEHV5-specific primers 7483/7485 (230-bp). Lanes 7 to 10, detection of EEHV6 in whole blood DNA samples collected on two successive days from African elephant #NAP35; lanes 7 to 8 EEHV PAN POL primers 6710/6711 followed by EEHV5-specific primer 7584/LGH6711 (470-bp); lanes 9 and 10; Codehops PAN-Herpesvirus POL primers 2595/2596 followed by primers 2596/2597 (250-bp). The center unmarked lane contains a 250-bp multimer size marker ladder.
3.9. Identification of EEHV6: a novel proboscivirus that is most closely related to EEHV1
Finally, evidence for yet another previously undetected Proboscivirus was obtained by PCR with the PAN EEHV primers on a blood sample collected from a 15-month old female African elephant calf with limb stiffness (a commonly encountered symptom in other EEHV cases). This animal was treated prophylactically with famciclovir and showed no further symptoms, but weak bands were obtained after first-round PCR and strong ones were sequenced after second-round PCR from two separate blood samples collected over two days at the time of diagnosis. The 250-bp second-round PCR POL DNA sequence obtained using either the PAN EEHV 6710/6711/6712 primer set (data not shown) or the PAN Herpesvirus Codehops 2595/2596/2597 primer set (Fig 4, lanes 9 and 10) again revealed a virus within the Proboscivirus genus, but this time with closest homology to EEHV1. A new specific inside primer 7584 was then designed based on this information and used together with primer 6711 to obtain the rest of the standard Codehops POL domain sequence as a 470-bp second-round PCR product (Fig 4, lanes 7 and 8). The combined 484-bp DNA sequence showed 40% G plus C content with an encoded DNA POL protein segment that differs from the EEHV1A, EEHV1B, EEHV5, EEHV2, EEHV3 and EEHV4 versions by 19, 20, 28, 30, 45 and 51 amino acids respectively (Table 3). Therefore, this new virus, which we have termed EEHV6, displays close to the same level of overall genetic divergence from EEHV1 as do EEHV5 from EEHV2 and EEHV4 from EEHV3. Because of the very young age of this calf and the fact that both Asian and African elephants were housed at this facility, we cannot make any interpretations as yet about whether EEHV6 is most likely to be endogenous to either African or Asian elephants. Clearly, it is also not known at this stage whether EEHV6 has the potential to cause severe pathogenesis in African elephants or not.
3.10. Phylogenetic comparison of all EEHV and EGHV DNA POL proteins
The sources, virus/case numbers and species identification of all eleven examples of elephant herpesviruses infections reported on and studied here are listed in the summary in Table 1, together with their Codehops region DNA POL sequence Genbank accession numbers. A predicted protein comparison over the 76-amino acid right-hand side of the POL Codehops region is shown in Figure 5 for all 13 known types of elephant herpesviruses, including the two sub-species pairs EEHV1A/1B and EGHV3A/3B.
Figure 5. Comparison of protein sequences within a 76 amino acid Codehops segment of the DNA POL genes of all 13 known elephant herpesvirus species or sub-species.
The predicted amino acids from codon positions equivalent to 702 to 777 of the EEHV1B (Kiba) DNA polymerase gene (Ehlers et al, 2006) are shown. The upper seven lines in the diagram represent the members of the Proboscivirus genus or EEHVs, whereas the lower six lines represent the elephant gammaherpesviruses or EGHVs. Conserved amino acids across both groups are shown in red, whereas amino acids that are specific to all EEHVs are shown in blue and those that are variable amongst the EEHVs are shown in black. Similarly, common amino acids that are unique to all of the elephant gammaherpesviruses are shown in grey and those that are unique to individual virus species within the elephant gammaherpesviruses are shown in black. Signature amino acids that are characteristic for EEHV1, EEHV4 and EGHV5 are overlined or underlined. Note that although EEHV1A and EEHV1B have an identical amino acid structure in this region of POL their DNA sequences differ at eight nucleotide positions here and several other proteins encoded by the chimeric EEHV1B genome differ from those of EEHV1A by between 8 and 40% at the amino acid level (Richman et al, unpublished data).
Protein level phylogenetic trees of the full 480-bp/160 amino acid Codehops DNA POL region obtained by the neighbor-joining procedure are presented together with bootstrap values in the dendrograms in Figure 6A and 6B. These show evolutionary distances and cladal branching patterns amongst each of the seven known EEHVs and six EGHVs, as well as comparisons with the orthologous DNA POL protein segments from many other selected beta and gammaherpesviruses from most other mammalian groups. New expanded data used here for the prototype strains of EEHV1A, EEHV1B and EEHV2 DNA POL Codehops regions was derived from Genbank Accession numbers GU350750, GU350752 and GU350753 (Stanton et al, 2010). Note that EEHV1A and EEHV1B do not differ at the protein level over the short Codehops region (Fig 5), although they exhibit 15 nucleotide and two amino acid changes over the longer Codehops region (Fig 6A), and display five common amino acid changes over a larger 1.2-kb segment of the DNA POL gene (data not shown).
Overall, the EGHVs proved to be diverged into three distinct subclasses (EGHV1/EGHV2, EGHV3A/EGHV3B/EGHV4 and EGHV5) that are widely scattered amongst the γ2-branch of the Gammaherpesvirinae and may eventually each rate separate genus status (Fig 6B). By contrast, the EEHVs all cluster within a single large clade defined collectively as the Proboscivirus genus representing a distinct β3-branch within the Betaherpesvirinae (Fig 6A). Although the cladal patterns of many of the deeper branches of the gammaherpesvirus tree lack robustness and change somewhat depending on whether nucleotide or protein comparisons are made, none of the EEHV or EGHV cladal patterns themselves were altered when an alternative method of phylogenetic analysis was used (UPGMA, data not shown). The Probosciviruses evidently separated from all other herpevirus groups very early in Betaherpesvirinae evolution and diverged into multiple subgroups with major branching events estimated to be of the order of 35, 20 and 10 million years ago.
4. Discussion
Until very recently, the three Probosciviruses EEHV1A, EEHV1B and EEHV2 that were identified originally as the causes of fatal elephant hemorrhagic disease (Richman et al., 1999, Fickel et al., 2003) were the only known elephant herpesviruses. Therefore, it was considered sufficient to use just the original PCR primer sets that were designed specifically for small segments of the TER or POL genes of EEHV1 and EEHV2 for all routine and diagnostic screening of samples from captive elephants in North America. This assay has been carried out successfully for confirming many new cases of EEHV disease at the first and/or second-round PCR level over the past decade within suspicious biopsies and emergency whole blood samples from moribund animals, as well as on occasion from relevant post-mortem necropsy samples. Importantly, numerous other blood, biopsy and necropsy samples that were submitted for routine screening evaluation proved to be negative.
However, that simplistic approach has now changed with the identification in mid-2007 of two cases of fatal hemorrhagic disease that were negative for both EEHV1 and EEHV2, but instead proved to contain two other distantly related viruses named EEHV3 and EEHV4 (Garner et al., 2009). Although the latter both still clearly belong to the Proboscivirus genus, they fall into to a new second G plus C-rich branch that is very highly diverged from the A plus T-rich branch containing EEHV1 and EEHV2 (Richman et al, unpublished data). After the initial detection of these new genomes with low efficiency redundant consensus TER or POL herpesvirus primers, we described a set of highly sensitive second-generation TER primers that are specific for EEHV3 plus EEHV4 and used them for preliminary genetic evaluation of these two novel viruses (Garner et al, 2009). However, the DNA POL gene locus is more variable than TER and is more commonly used in pathological studies. It also offers a more precise and reliable indicator of phylogenetic relationships among herpesviruses. Therefore, we needed to also develop a matching set of EEHV3 and EEHV4-specific POL primers as described here.
In addition, to encompass the possibility of there being more as yet undiscovered elephant herpesviruses both within the Proboscivirus genus (EEHVs) and amongst the newly described elephant gammaherpesviruses or EGHVs (Wellehan et al., 2008), we also developed two sets of moderately redundant POL locus PCR primers focused on broad range detection of each of these two classes of elephant herpesviruses. Our rationalization that these would likely be valuable diagnostic reagents with regard to understanding and controlling elephant herpesvirus-associated disease has been amply confirmed by the subsequent rapid identification by PCR sequencing procedures of eleven more examples of elephant herpesviruses (other than EEHV1) within blood, lesions or nodules of healthy and diseased Asian and African elephants. These include PCR DNA sequence based evidence for novel fifth and sixth distinct species of the Probosciviruses (EEHV5 and EEHV6), as well as for both a new fifth species of elephant gammaherpesvirus (EGHV5) and a significantly diverged variant EGHV3B of the previously described EGHV3A virus.
For human HCMV and EBV infections, the highest viral load levels detected in blood are in the range of 105 to 106 viral genomes per ml, and for both viruses, transplant or AIDS patients are considered to have systemic viremia requiring antiviral medication at between 5 × 103 and 1 × 104 viral genomes per ml. Therefore, considering that during acute disease the EEHVs attack and damage vascular endothelial cells, our semi-quantitative estimates of viral loads for EEHV3 and EEHV4 being 107 viral genome copies per ml or greater, make a compelling case that these two viruses were indeed the primary etiological agents responsible for the hemorrhagic endothelial cell disease and deaths of these two juvenile Asian elephants. We emphasize that our original detection of EEHV3 and EEHV4 with the standard PAN mammalian herpesvirus PCR primers was highly serendipitous and was successful only because of the extraordinarily high viral load in just one tissue sample from each case. These viruses have proven to be so diverged in their DNA POL sequences that they each have four to five nucleotide mismatches with one or more of the highly redundant generic primers. In fact, both the standard PAN-herpes TER and POL redundant primers all gave negative results on first and second-round PCR in the NAP#27 and NAP#22 blood samples, despite the high viral load there, compared to with the later EEHV3/4-specific primers, which improved the sensitivity by two orders of magnitude.
Wellehan et al (2008) described four distinct species of gammaherpesviruses detected by PCR in conjunctival or genital swabs from 11 of 21 tested healthy elephants in captivity. These included three sequenced POL DNA fragments referred to by them as ElHV3, ElHV4 and ElHV5 in Asian elephants and another ElHV6 in the single African elephant tested. Some of the positive Asian elephant swabs were taken from lymphoid vulvitis conditions that have been described as being very common in both Asian and African elephants (Munson, 1995). To clearly distinguish this class of elephant herpesviruses from the Probosciviruses, we and others (Ehlers et al., 2008) have proposed to instead name and number them as elephant gammaherpesviruses (eg EGHV1, EGHV2, EGHV3 and EGHV4 respectively). The four new Probosciviruses EEHV3, EEHV4, EEHV5 and EEHV6 as well as the new fifth elephant gammaherpesvirus EGHV5 now represent the seventh, eighth, ninth, tenth and eleventh distinctive species of herpesviruses found among either Asian or African elephants, which would likely be named ElHV7 to ElHV11 under the current formal ICTV nomenclature (Table 4).
More extensive genetic analysis to be reported elsewhere (Richman et al, unpublished data) confirms that all six Probosciviruses differ significantly at multiple additional genetic loci, such as the U73/Origin Binding Protein and U77/Helicase genes as well as in the U38/DNA POL gene, and that EEHV3 and EEHV4, EEHV2 and EEHV5, and EEHV1 and EEHV6 tend to cluster together into three paired sister clades in phylogenetic trees at all of these loci. Furthermore, over a larger scale, the EEHV1 and EEHV2 genomes, whilst essentially colinear, diverge by an average of 20% at the protein level over all 30 gene loci examined across an 85-kb central segment of their expected to be 200-kb genomes. In addition, the EEHV1 viral genomes are themselves divisible into two distinct subtypes called EEHV1A and EEHV1B that are complex chimeric mosaics relative to one another. Including the EGHV3A/EGHV3B variant pair described here, the EEHV1A/EEHV1B pair makes a total of thirteen distinct types of herpesviruses altogether that are now known to infect Asian and/or African elephants (Table 4). As illustrated in these studies, maximal efficiency and accurate detection of these numerous elephant herpesviruses when screening pathological samples requires the use of a complex series of both partially redundant generic and highly-specific non-redundant PCR primer sets, as well as direct DNA sequencing of all PCR products obtained.
Infections with the EGHVs within captive elephant populations have apparently remained ubiquitous and largely asymptomatic. In general, gammaherpesviruses are thought to persist at a low level in the latent state primarily in lymphoid cells, although their frequent detection in genital mucosal and conjunctival secretions of elephants as described by Wellehan et al (2008) implies that reactivation may be common in these hosts. We estimate that the levels of EGHV2 (#NAG1) DNA present in the blood of Case #NAP27 were at least 5,000-fold lower than the very high levels of EEHV3 (#NAP27) DNA carried there. Nevertheless, this result represents the first example of detection of any elephant gammaherpesvirus associated with systemic infection within blood or internal organs, where it was most likely a reactivated passenger accompanying a primary infection by the pathogenic EEHV3. Although no EGHV2 was detected within the acute disease tissue or blood samples from Case #NAP22, our experiments did not address the possibility of other EGHVs being present at similar low levels.
So far, neither the EEHV1A, EEHV2, EEHV3 nor EEHV4 viruses have ever been found at high enough levels for diagnostic first or second-round PCR detection within routine blood samples from any of the numerous healthy captive Asian elephants that have undergone testing, nor in unrelated necropsy tissue samples. Rather (with a single recent exception involving EEHV1B) they have all only been found in association with cases of acute systemic disease, which all included a high level of viremia as well as microvascular endothelial cell damage in most major organs in necropsy tissue (Richman et al., 2000a, Garner et al., 2009). However, new tests with the more sensitive second-generation primer sets and by real-time PCR have begun to detect low levels of virus in blood at early stages of viremia as well as in trunk washes of healthy herdmates, suggesting that (like the EGHVs) latent infection and very low level localized reactivation and shedding by EEHV1 at least may also be quite common (Stanton et al., 2010).
The clearly implied causal association of EGHV5 with a proliferating mucosal epithelial papilloma also represents a novel finding and should generate caution about automatically assuming that all such abnormal growths, including those containing inclusion bodies, are necessarily associated with the pathogenic EEHVs. On the other hand, the two examples of both EGHV2 (#NAG2 and #NAG3) and EGHV3 (#NAG4 and #NAG5) detected in biopsied vulval lesions or blood of otherwise asymptomatic captive elephants may be similar to the observations using genital secretion swabs by Wellehan et al (2008), and do not necessarily imply that they were directly causative of the biopsied lesions themselves. Furthermore, one of our two cases with genital lesions carrying both EEHV2 and EEHV3 was an African elephant. Both of these two virus species were recognized only within Asian elephant swab samples by Wellehan et al (2008), but our wild-born African sample contained significantly diverged versions of both viruses, especially that of EGHV3B, which we judge to be sufficiently different to warrant being named as a distinctive variant compared to EGHV3A. Although this a complex issue, the differences found between the African versus Asian elephant versions of EGHV2 and EGHV3 may imply that there has not (yet) been much intermingling of elephant host-specific versions of the gammaherpesviruses in captivity. It also raises the question of whether there may well be additional distinct African and Asian elephant versions of some of the other gammaherpesviruses and even of the Probosciviruses that have not yet been identified.
In contrast to the EGHVs, infections with EEHVs can sometimes be highly pathogenic, especially in juvenile Elephas maximus. EEHV1A, EEHV1B, EEHV2, EEHV3 and EEHV4, but not yet EEHV5 or EEHV6, have all been found associated with cases of fatal hemorrhagic disease. The phylogenetic trees show that the Probosciviruses have evidently co-evolved with their natural hosts the Elephantids throughout the past 100 million years since they separated from all other mammalian groups. Therefore, infections with these viruses would be expected to be well-adapted to and universal in modern elephants, as well as capable of persisting in a long-term latent state in the wild within their natural host species. However, instead of the normal asymptomatic primary infections as infants, something as yet unknown about the situation (with EEHV1 especially) in captive juvenile Asian elephants leads to an inability of the victims to cope with or control what we presume to be primary infections. The key factors involved here could include some or all of the following: (1) Far later than normal primary infections; (2) The absence of maternal antibodies or of potentially protective latent infections with other members of the EEHV group; and (3) Being infected with the wrong species or sub-species of EEHV, which have not become evolutionarily adapted to a particular Asian elephant host species or subspecies. This latter situation may parallel that of the Rhesus B-virus (Macaquine herpesvirus 1), an alphaherpesvirus that is asymptomatic in its natural host species, but has high lethality in the rare occasions in which it is transmitted to humans (Elmore and Eberle, 2008).
Other than the sometimes successful treatment with the anti-herpesvirus drugs famciclovir or ganciclovir (Schmitt et al., 2000), the best long-term solution to this devastating disease will most likely be the development of adapted attenuated live or killed EEHV vaccines. However, the inability as yet to be able to grow any of these viruses in cell culture, together with the now very large number of different herpesviruses identified, greatly complicates the situation. Attempts to obtain additional genomic DNA sequence data for more robust comparative phylogenetic analyses between the most closely related Proboscivirus pairs EEHV3 and EEHV4, or EEHV2 and EEHV5, as well as EEHV1 and EEHV6 are in progress. The PCR reagents described here should make possible the detection of both additional virus species and as yet unidentified cases of this disease, as well as set the stage for efforts to try to definitively deduce the natural source host species for each of the pathogenic Probosciviruses.
5. Conclusions
Six new PCR primer sets were designed to be: (1) Specific for the EEHV3 plus EEHV4 POL gene; (2) Generic for all known EEHV POL genes; (3) Generic for all known EGHV POL genes; (4) Specific for EEHV5 and (5) Specific for EEHV6. In addition to being used to diagnose or confirm several additional new cases of EEHV1-associated acute disease (to be addressed elsewhere), the generic primer sets were effective in detecting numerous examples of elephant herpesviruses other than EEHV1A or EEHV1B in either routine or pathological samples from Asian and African elephants. Based on initial DNA sequence data from the generic primers, we then generated and tested more sensitive specific primers for each of the new viruses and used them to evaluate virus loads and extend the genetic comparisons of all of these species over the full 480-bp Codehops segment of the DNA polymerase PCR locus.
Specifically, we have shown here that: (a) The new EEHV3/4 POL primers are capable of detecting both EEHV3 and EEHV4 in all necropsy tissue samples of the two lethal cases; (b) There were extremely high levels of viremia reaching at least 107 viral genomes per ml in the blood of the lethal EEHV3 and EEHV4 cases; (c) Low levels of the known elephant gammaherpesvirus EGHV2 and EGHV3 were detected in blood from two Asian elephants, one being the fatal case of EEHV3 disease and the other having oral lesions; (d) Coinfection with both EGHV2 and EGHV3 was detected in two elephants with vulval lesions, in one case from the blood of an Asian elephant and in the second case directly within a biopsy from an African elephant; (e) The PCR DNA sequence data here revealed apparent Asian (EGHV3A) and African (EGHV3B) specific variants of EGHV3; (f) A fifth elephant gammaherpesvirus named EGHV5 was identified in a mucosal trunk papilloma biopsy from a healthy captive Asian elephant; (g) A fifth distinct Proboscivirus named EEHV5 was also identified in routine blood samples from a healthy adult Asian elephant; and finally (h) Another novel Proboscivirus named EEHV6 was identified in blood from an ailing African elephant calf.
Clearly, both Asian and African elephants naturally harbor multiple species of herpesvirusus from at least two distinct sub-families: the Proboscivirus genus within the betaherpesviruses and and a diverged group of gammaherpesviruses. As expected, these viruses have complex interactions with their elephant hosts, which may include potential cross-species infections, and unfortunately with the EEHVs this can lead to a lethal hemorrhagic disease in calves that is currently a major worldwide problem for successful breeding programs for endangered Asian elephants. Extensive surveys to search for other potential examples of EEHV3, EEHV4, EEHV5, EEHV6 or EGHV5 within captive or wild Asian and African elephants would likely be highly informative.
Acknowledgements
The authors wish to acknowledge the assistance of all of the keepers, curators, veterinarians and healthcare staff who forwarded samples to the National Elephant Herpesvirus Laboratory for evaluation. We are especially indebted to the following facilities that contributed directly to this study, including the Woodland Park Zoo, Seattle, WA; the Carson and Barnes Elephant Facility, Hugo, OK; the Ringling Bros. and Barnum and Bailey Center for Elephant Conservation, Orlando, FL; Knoxville Zoo, Tennessee; Columbus Zoo, OH; Riddles Elephant and Wildlife Sanctuary, AR; Houston Zoo, TX and the Smithsonian's National Zoological Park in Washington, DC. We thank James Wellehan (University of Florida) for initially alerting us to the fact that Case #NAP27 was also infected with an elephant gammaherpesvirus and Mike Garner (Northwest ZooPath, Monroe, WA) for providing the trunk papilloma sample from Case #NAG6.
Studies at the National Zoological Park in Washington DC were funded by Research grants to LKR from the Smithsonian Institution, the Morris Animal Foundation, the International Elephant Foundation, and Ringling Bros. and Barnum and Bailey Center for Elephant Conservation. Studies at the Johns Hopkins School of Medicine were funded through NIH research grant R01 AI24576 to GSH. Protocols for collection of elephant clinical samples were reviewed and approved by the both local and/or National Zoological Park IACUC. The financial sponsors had no involvement in the experimental design, analysis or interpretation of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Burkhardt S, Goltz M, Bergmann V, Ochs A, Weiler H, Hentschke J. Genetic and ultrastructural characterization of a European isolate of the fatal endotheliotropic elephant herpesvirus. J Gen Virol. 2001;82:475–482. doi: 10.1099/0022-1317-82-3-475. [DOI] [PubMed] [Google Scholar]
- Ehlers B, Dural G, Marschall M, Schregel V, Goltz M, Hentschke J. Endotheliotropic elephant herpesvirus, the first betaherpesvirus with a thymidine kinase gene. J Gen Virol. 2006;87:2781–2789. doi: 10.1099/vir.0.81977-0. [DOI] [PubMed] [Google Scholar]
- Ehlers B, Dural G, Yasmum N, Lembo T, de Thoisy B, Ryser-Degiorgis MP, Ulrich RG, McGeoch DJ. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. Journal of virology. 2008;82:3509–3516. doi: 10.1128/JVI.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore D, Eberle R. Monkey B virus (Cercopithecine herpesvirus 1) Comp Med. 2008;58:11–21. [PMC free article] [PubMed] [Google Scholar]
- Fickel J, Lieckfeldt D, Richman LK, Streich WJ, Hildebrandt TB, Pitra C. Comparison of glycoprotein B (gB) variants of the elephant endotheliotropic herpesvirus (EEHV) isolated from Asian elephants (Elephas maximus) Vet Microb. 2003;91:11–21. doi: 10.1016/s0378-1135(02)00264-x. [DOI] [PubMed] [Google Scholar]
- Fickel J, Richman LK, Montali R, Schaftenaar W, Goritz F, Hildebrandt TB, Pitra C. A variant of the endotheliotropic herpesvirus in Asian elephants (Elephas maximus) in European zoos. Vet Microb. 2001;82:103–109. doi: 10.1016/s0378-1135(01)00363-7. [DOI] [PubMed] [Google Scholar]
- Garner MM, Helmick K, Ochsenreiter J, Richman LK, Latimer E, Wise AG, Maes RK, Kiupel M, Nordhausen RW, Zong JC, Hayward GS. Clinicopathologic features of fatal disease attributed to new variants of endotheliotropic herpesviruses in two Asian elephants (Elephas maximus) Vet Pathol. 2009;46:97–104. doi: 10.1354/vp.46-1-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson E, Sundberg J, Gaskin J, Kollias G, O'Banion M. Cutaneous papillomas associated with a herpesvirus-like infection in a herd of captive African elephants. J Am Vet Med Assoc. 1986;189:1075–1078. [PubMed] [Google Scholar]
- McCully RM, Basson PA, Pienaar JG, Erasmus BJ, Young E. Herpes nodules in the lung of the African elephant (Loxodonta africana (Blumebach, 1792)) Onderstepoort J Vet Res. 1971;38:225–235. [PubMed] [Google Scholar]
- Munson L. Lymphoid follicular vulvitis in African (Loxodonta Africana) and Asian (Elephans maximus)elephants. J. Zoo Wildl Med. 1995;26:353–358. [Google Scholar]
- Ossent P, Guscetti F, Metzler A, Lang E, Rübel A, Hauser B. Acute and fatal herpesvirus infection in a young Asian elephant (Elephas maximus) Vet Pathol. 1990;27:131–133. doi: 10.1177/030098589002700212. [DOI] [PubMed] [Google Scholar]
- Reid C, Hildebrandt T, Marx N, Hunt M, Thy N, Reynes J, Schaftenaar W, Fickel J. Endotheliotropic elephant herpes virus (EEHV) infection. The first PCR-confirmed fatal case in Asia. Vet Q. 2006;28:61–64. doi: 10.1080/01652176.2006.9695209. [DOI] [PubMed] [Google Scholar]
- Richman L, Montali R, Cambre R, Schmitt D, Hardy D, Hildbrandt T, Bengis R, Hamzeh F, Shahkolahi A, Hayward G. Clinical and pathological findings of a newly recognized disease of elephants caused by endotheliotropic herpesviruses. J Wildl Dis. 2000a;36:1–12. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- Richman L, Montali RJ, Hayward GS. Review of a Newly Recognized Disease of Elephants Caused by Endotheliotrophic Herpesviruses. Zoo Biol. 2000b;19:383–392. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. Novel Endotheliotropic Herpesviruses Fatal for Asian and African Elephants. Science. 1999;283:1171–1176. doi: 10.1126/science.283.5405.1171. [DOI] [PubMed] [Google Scholar]
- Rose TM. CODEHOP-mediated PCR - a powerful technique for the identification and characterization of viral genomes. Virology journal. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt D, Hardy D, Montali R, Richman L, Lindsay W, Isaza R, West G. Use of famciclovir for the treatment of endotheliotrophic herpesvirus infections in Asian elephants (Elephas maximus) J Zoo Wildl Med. 2000;31:518–522. doi: 10.1638/1042-7260(2000)031[0518:UOFFTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Stanton JJ, Zong J-C, Latimer E, Tan J, Herron A, Hayward GS, Ling PD. Detection of Pathogenic Elephant Endotheliotropic Herpesvirus in Routine Trunk Washes from Healthy Adult Asian Elephants (Elephas maximus) using a Novel Quantitative Real-Time PCR Assay. Am. J. Vet Res. 2010 doi: 10.2460/ajvr.71.8.925. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellehan JF, Johnson AJ, Childress AL, Harr KE, Isaza R. Six novel gammaherpesviruses of Afrotheria provide insight into the early divergence of the Gammaherpesvirinae. Vet Microbiol. 2008;127:249–257. doi: 10.1016/j.vetmic.2007.08.024. [DOI] [PubMed] [Google Scholar]