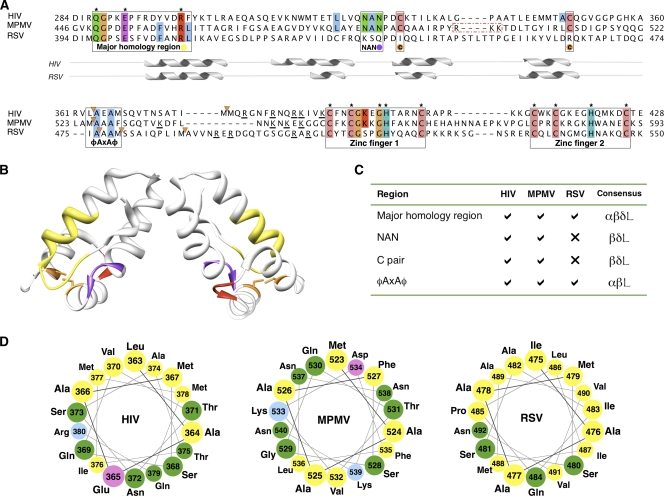
FIG. 3.
Comparisons of the Gag sequences from HIV, M-PMV, and RSV. (A) Multiple alignment of Gag C-CA-NC for RSV, M-PMV, and HIV. Regions of interest are boxed, and colored circles indicate the coloring of these regions in panels B and C. Residues conserved across >75% of viruses in an extended alignment, including 12 viral species (see Fig. S3 in the supplemental material), are colored using ClustalX coloring; residues conserved across all species are specified by an asterisk. Orange triangles denote cleavage sites. Basic residues adjacent to the NC are underlined. A dashed red box surrounds an RKK motif unique to M-PMV C-CA. The position of helices from the solved structures for RSV (PDB: 1d1d) and HIV (PDB: 3ds2) are shown under the alignment for illustrative purposes. Residues are numbered from the beginning of Gag. (B) HIV C-CA (PDB: 3ds2) with residues colored to indicate the positions of regions of interest: yellow, MHR; purple, NAN; orange, paired cysteine residues; and red, RKK motif (see the text and panels A and C). (C) Table of regions of interest defined in panel A; the colors correspond to the coloring in the multiple alignment (A) and HIV structure (A). Consensus signifies genera in which the feature is found (α signifies alpha, β signifies beta, δ signifies delta, and L signifies lentiretroviruses). There is currently no solved structure for the CA region in M-PMV, the final helix of M-PMV is defined assuming structural homology to HIV (PDB: 3ds2). (D) Helical wheel representation of the 18 residues after the CA adjacent cleavage site for the proteins of interest.