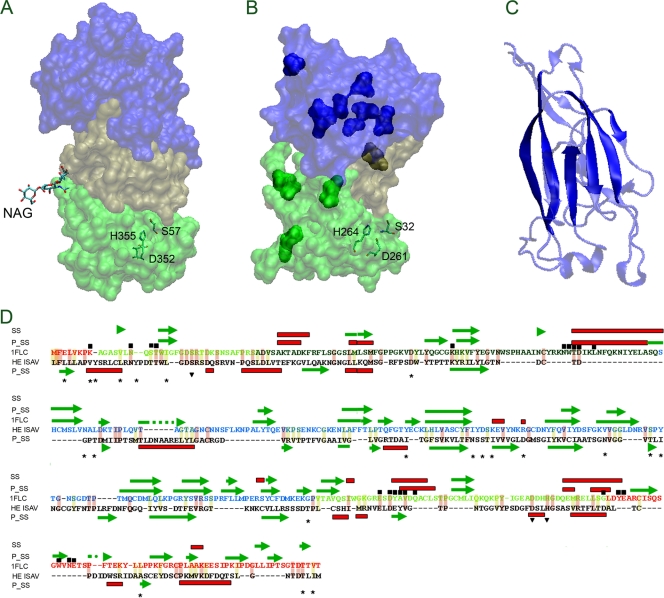
FIG. 4.
Structural characterization of the HE protein from ISAV by homology modeling. (A) Structure of HEF from influenza C virus (PDB ID 1FLC); (B) structural model of HE from ISAV. Their characteristic subdomains E1 and E2, R, and E′ are indicated in light green, blue, and dark green, respectively. The catalytic residues of HAF from influenza C virus (S57, D352, and H355) and putative catalytic residues from ISAV (H264, D261, and S32) are indicated. (C) Structure of the R domain from influenza C virus. The solid colors indicate conserved structural elements with the predicted secondary structure of HE from ISAV. (D) Alignment between influenza C virus HEF and ISAV HE using secondary structural prediction (using Phyre). The figure shows the predicted secondary structure (P_SS), experimental secondary structure (SS), the catalytic residues (residues over the inverted triangle), residues close to (6 A) NAG (residues under the black box), the variable position among HE proteins coming from ISA virus with its entire genome sequenced (residues over the stars). The helixes are shows in red boxes and β-strands in green arrows. The conserved changes are indicated with light yellow background and the identical residues with light red background. The secondary element b-strand (green arrow) and helix (red box) are also indicated.