Fig. 1.
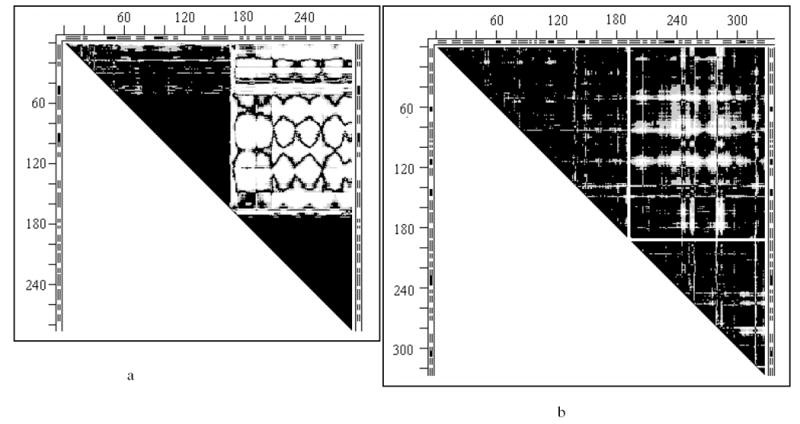
Distance difference matrices (DDMs)
- 2tbvAC for the molecules A and C in the unit cell of the 2tbv virus capsid protein; the mostly white rectangle at the DDM’s top right, providing straight white borders of the “rigid body triangles” of this DDM, indicates that most DDs between the two “rigid bodies” are larger than 1Å, meaning that most Cα-Cα distances between the “rigid bodies” differ in two molecules by more than 1Å.
- 4ape5er2 of the apo-holo structures of endothiapepsin; this DDM has only some white spots and no straight white borders of the black “rigid triangles”; such borders can be provided by drawing white lines encompassing almost all white spots in a rectangle and thus delineating the mostly black “rigid” triangles (see also text on thermolysin below).
