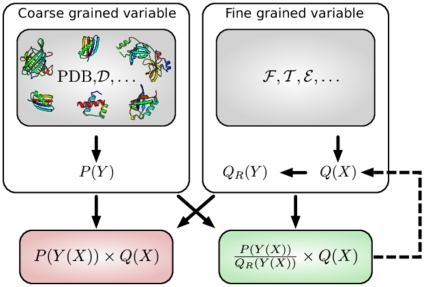
Figure 2. General statistical justification of PMFs.
The goal is to combine a distribution  over a fine grained variable
over a fine grained variable 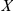 (top right), with a probability distribution
(top right), with a probability distribution 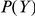 over a coarse grained variable
over a coarse grained variable  (top left).
(top left).  could be, for example, embodied in a fragment library (
could be, for example, embodied in a fragment library ( ), a probabilistic model of local structure (
), a probabilistic model of local structure ( ) or an energy function (
) or an energy function ( );
);  could be, for example, the radius of gyration, the hydrogen bond network, or the set of pairwise distances.
could be, for example, the radius of gyration, the hydrogen bond network, or the set of pairwise distances.  usually reflects the distribution of
usually reflects the distribution of  in known protein structures (PDB), but could also stem from experimental data (
in known protein structures (PDB), but could also stem from experimental data (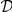 ). Sampling from
). Sampling from  results in a distribution
results in a distribution  that differs from
that differs from  . Multiplying
. Multiplying 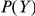 and
and  does not result in the desired distribution for
does not result in the desired distribution for  either (red box); the correct result requires dividing out the signal with respect to
either (red box); the correct result requires dividing out the signal with respect to  due to
due to  (green box). The reference distribution
(green box). The reference distribution 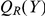 in the denominator corresponds to the contribution of the reference state in a PMF. If
in the denominator corresponds to the contribution of the reference state in a PMF. If 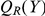 is only approximately known, the method can be applied iteratively (dashed arrow). In that case, one attempts to iteratively sculpt an energy funnel. The procedure is statistically rigorous provided
is only approximately known, the method can be applied iteratively (dashed arrow). In that case, one attempts to iteratively sculpt an energy funnel. The procedure is statistically rigorous provided  and
and  are proper probability distributions; this is usually not the case for conventional pairwise distance PMFs.
are proper probability distributions; this is usually not the case for conventional pairwise distance PMFs.