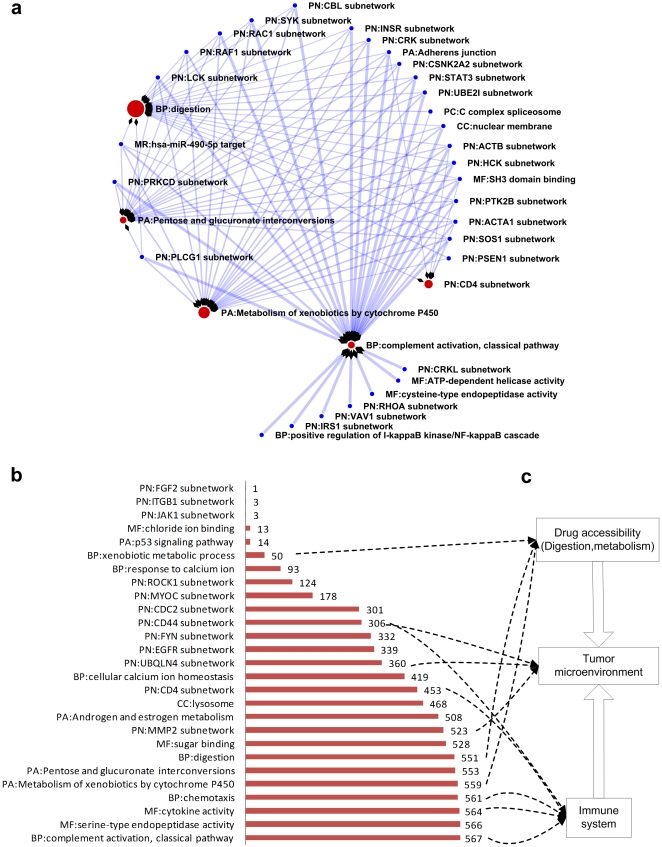
Figure 2. Topological characteristics of the Pattern of Action network.
a. Gatekeeper modules and checkpoint modules, demonstrated by an example (the POA result of Cisplatin on non-small cell lung carcinoma). We define the flow-out nodes (blue circles) as ‘checkpoint modules’ (from gene signatures of drug sensitivity), and the flow-in nodes (red circles) as ‘gatekeeper modules’ (which cooperate with a large number of modules to determine the clinical prognosis outcome). The radius of the red circles is proportional to the in-degree (number of flow-in links) of the node in the generic inter-module cooperation network. b. All of the ‘gatekeeper’ modules in a generic inter-module cooperation network generated for lung cancer (non-small cell lung carcinoma). The length of bars and annotated numbers indicate in-degree (number of flow-in links) for each gatekeeper module (y-axis). c. An ensemble of common gatekeeper modules in multiple cancer types highlights a physiology-level ‘pathway’ of drug action. Gene module names start with a 2-character header that indicates the gene module definition source, PN: protein subnetwork; PA: pathway; BP: Gene Ontology biological process; MF: Gene Ontology molecular function; CC: Gene Ontology cellular component; MR: microRNA targets.