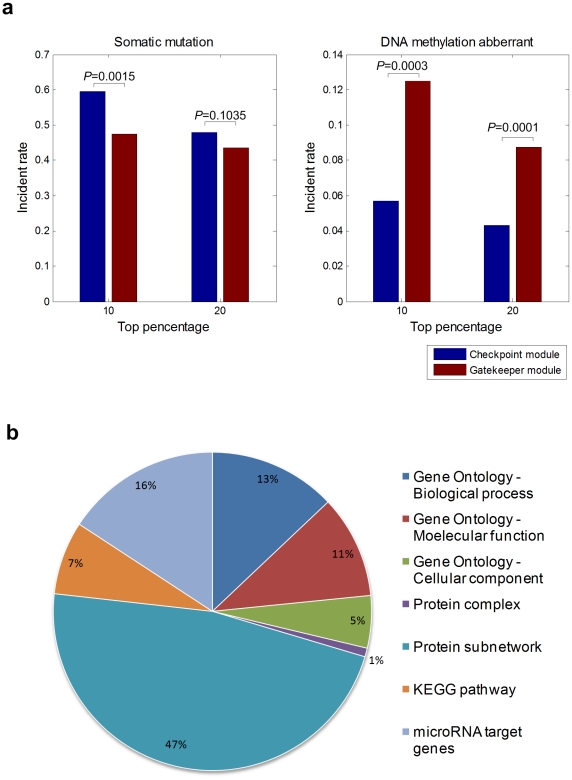
Figure 3. Biological function characterization of the inter-module cooperation network.
a. For each type of module (gatekeeper and checkpoint), the top 10% and 20% most highly used genes are used as the representative genes for each module, and their incident rate of somatic mutation frequency and DNA methylation aberrant was calculated for lung cancer (NSCLC); p-value for incident rate difference was calculated using the binomial distribution (see Methods ). b. Contribution of various evidence sources for gene module definition in lung cancer (NSCLC). We summarized the number of various types of gene module definitions in the identified inter-module network for lung cancer (NSCLC) and the proportional contribution of various evidence sources for the gene modules were plotted.