Abstract
Background
Well-curated and validated network reconstructions are extremely valuable tools in systems biology. Detailed metabolic reconstructions of mammals have recently emerged, including human reconstructions. They raise the question if the various successful applications of microbial reconstructions can be replicated in complex organisms.
Results
We mapped the published, detailed reconstruction of human metabolism (Recon 1) to other mammals. By searching for genes homologous to Recon 1 genes within mammalian genomes, we were able to create draft metabolic reconstructions of five mammals, including the mouse. Each draft reconstruction was created in compartmentalized and non-compartmentalized version via two different approaches. Using gap-filling algorithms, we were able to produce all cellular components with three out of four versions of the mouse metabolic reconstruction. We finalized a functional model by iterative testing until it passed a predefined set of 260 validation tests. The reconstruction is the largest, most comprehensive mouse reconstruction to-date, accounting for 1,415 genes coding for 2,212 gene-associated reactions and 1,514 non-gene-associated reactions.
We tested the mouse model for phenotype prediction capabilities. The majority of predicted essential genes were also essential in vivo. However, our non-tissue specific model was unable to predict gene essentiality for many of the metabolic genes shown to be essential in vivo. Our knockout simulation of the lipoprotein lipase gene correlated well with experimental results, suggesting that softer phenotypes can also be simulated.
Conclusions
We have created a high-quality mouse genome-scale metabolic reconstruction, iMM1415 (Mus Musculus, 1415 genes). We demonstrate that the mouse model can be used to perform phenotype simulations, similar to models of microbe metabolism. Since the mouse is an important experimental organism, this model should become an essential tool for studying metabolic phenotypes in mice, including outcomes from drug screening.
Background
The first genome-scale reconstruction of metabolic networks emerged eleven years ago [1], four years after the first whole genome sequencing of an entire organism was published [2]. To date, 29 bacteria, 2 archaea and 5 eukaryotes have been reconstructed and for some organisms, up to 5 updates have been published [3]. The reconstruction process is well established for metabolic networks [4]. Once assembled, the reconstruction can be readily converted into a mathematical format by adding balances (e.g., mass -balance constraints), steady-state assumptions and bounds (e.g. physical constraints) [5]. The resulting model is condition-specific and can be used for phenotype simulations using various constraint-based reconstruction and analysis (COBRA) methods [5,6]. This approach has proven successful for various microorganisms and eukaryotes for addressing various biological and biotechnological questions, such as the analysis of knowledge gaps [7], simulation of phenotype traits [8], analysis of evolution of metabolic networks [9,10] and metabolic engineering applications [11]. The numerous applications have recently been reviewed [3,12].
The release of the human genome [13] and its annotation has provided the appropriate foundation for human metabolic reconstructions. Three approaches have been published to date. Two of those, the HumanCyc [14] and the Edinburgh Human Metabolic Network [15,16] were created with a largely automated top-down approach, while Recon 1 [17] was created by an extensive bottom-up manual curation of an automated draft map, including extensive gap-assessment and filling using bibliomic data. Recon 1 has eight compartments (cytoplasm, extracellular space, mitochondria, Golgi apparatus, endoplasmic reticulum, lysosome, peroxisome and nucleus) and accounts for 1496 genes coding for 2004 proteins [17]. Overall, Recon 1 covers all major metabolic pathways occurring in any human cell by accounting for 2,233 biochemical transformations and 1,510 transport and exchange reactions. The Recon 1 can be converted into a functional in silico model can be converted into a functional model of human metabolism, which was validated by applying 288 flux balance analysis (FBA) based tests of core functionality of human metabolism [17]. A recent use of Recon 1 includes a study on the topology of human disease finding that several co-morbid diseases had a functional relationship within Recon 1 [18]. Also, expression profiling data mapped onto Recon 1 was used to create a cellular network for ten human tissue types [19]. Furthermore a systems based analysis of the effects of imprinted genes on metabolic functions has recently been published [20].
Metabolic networks have been reconstructed for two further mammals, a central metabolic reconstruction of cattle (Bos taurus) [21] and four mouse (Mus musculus) reconstructions [22-25].Two of these mouse reconstructions were constructed manually [24,25], while the remaining two were generated semi-automatically [22,23]. The first genome-scale manual curated reconstruction represents the metabolism of mouse hybridoma cell lines [25] and has recently been expanded to represent 1494 reactions coded by 724 genes in three compartments [24]. In addition, a pre-genome sequencing reaction network of intermediate metabolism in mouse hybridoma cell lines was built and analyzed using linear optimization [26].
The mouse serves as a fundamental experimental animal for human biomedical applications. Furthermore, the availability of phenotyped inbred knockout mouse strains [27] make it ideal for examining the phenotype prediction capabilities of mammalian reconstructions. Therefore, a detailed reconstruction of mouse metabolism that can be converted into highly functional model is of a great importance.
Given the high sequence homology between most mammalian genomes, an obvious starting point for metabolic reconstruction of mammals is the comprehensive human metabolic reconstruction, Recon 1. We therefore sought to create draft mammalian metabolic reconstructions based on Recon 1. The mouse reconstruction was then manually completed and validated. We also present simulation results using a functional model of mouse metabolism to test its phenotype prediction capabilities.
Results
Identification of homologous genes
The HomoloGene database contains information on homologous genes in 5 mammals and 15 non-mammals [28]. We searched the HomoloGene database for all 1496 genes of Recon 1 and found a human match for 1,464 genes (97.8%). The mammalian organism with the highest number of genes homologous to Recon 1 genes was the mouse (Mus musculus) (1,415 genes, 97%). The non-mammalian organism with the highest number of genes homologous to Recon 1 genes was the zebra-fish (Danio rerio) (1,200 genes, 82%) (Additional file 1).
Creation of draft metabolic network reconstructions
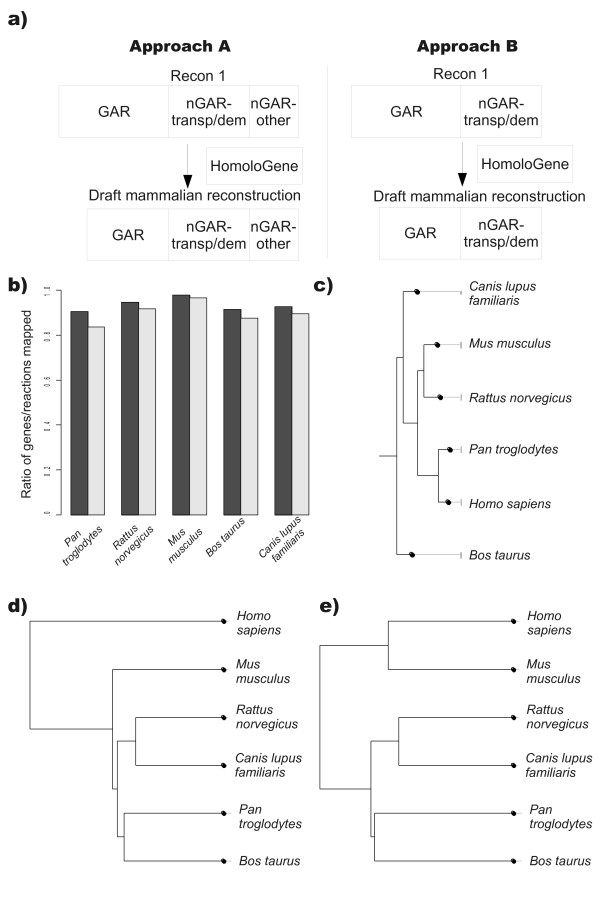
For each mammal, a draft reconstruction was created via two different approaches (Figure 1a). In approach A (modelA), all reactions linked to Recon 1 genes that did not have a homologous gene were removed from the reconstruction of the corresponding species. Therefore, modelA included all reactions linked to genes homologous to Recon 1 genes in addition to all non-gene associated reactions (1,514). Of the non-gene-associated reactions, 676 were transporter reactions, 452 were demand or exchange reactions, leaving 385 reactions within a metabolic pathway but without gene-association. In approach B (modelB), we included only genes and their reactions homologues to Recon 1 genes as well as transporter and demand reactions. All other reactions, including non-gene associated reactions, were removed from the reconstruction.
Figure 1.
Creation of draft mammalian reconstructions. a) A schematic figure showing the two approaches used to generate draft mammalian reconstructions using Recon 1. Approach A removes all gene-associated reactions from Recon 1 without a homologous gene in the reconstructed animal, while keeping all non-gene-associated reactions. Approach B removes all gene-associated reactions from Recon 1 without a homologous gene in the reconstructed as well as non-gene-associated reactions (excluding transporters and demand reactions). GAR - gene-associated-reactions; nGAR - non-gene-associated reaction; transp/dem - transporters and demand reactions. b) Ratio of reactions (black bar) and genes (gray bar) that were successfully mapped from Recon 1 to the indicated mammalian draft reconstruction. c) A phylogenetic tree based on all transcripts of protein domain sequences from the SuperFamily database [64] for all reconstructed mammals. d) A phylogenetic tree based on flux variability analysis (FVA) of all reactions in all mammals reconstructed via approach A. e) A phylogenetic tree based on flux variability analysis (FVA) of all reactions in all mammals reconstructed via approach B.
The ratio of Recon 1 genes with a homologous gene as well as the ratio of reactions from Recon 1 included in the draft reconstructions for each mammal is shown in Figure 1b. The mammal with the highest ratio of mapped genes (97%) and reactions (98%) was the mouse while the chimpanzee (Pan troglodytes) had the lowest ratio of both mapped genes (84%) and reactions (91%). Furthermore, we created for each mammalian modelA and modelB a compartmentalized and non-compartmentalized version. The more complex, compartmentalized versions had more metabolic dead ends (321 vs. 150) and therefore more reactions with zero fluxes. The number of reactions with non-zero fluxes for both compartmentalized and non-compartmentalized versions of all draft mammalian models created using approach A and B are summarized in Table 1. We performed flux variability analysis (FVA) of models of these reconstructions, allowing uptake of all extracellular metabolites in all models in order to maximize likelihood of non-zero fluxes. Compared to humans, the mouse model had the highest percentage (84-89%) and the chimpanzee model had the lowest percentage (41-54%) of active reactions with non-zero fluxes for all model versions (Table 1). We then constructed a phylogenetic tree of the reconstructed mammals based on the FVA results (Figure 1d,e). The results indicated that the mouse draft model had the highest degree of similarity with the human model for both modelA (Figure 1d) and modelB (Figure 1e). These findings are surprising as the chimpanzee is the closest relative of humans with approximately 99% overall genome similarity [29]. It is also the closest ancestor to humans of the mammals reconstructed when protein coding sequence is compared (Figure 1c).
Table 1.
Results from flux variability analysis of the draft mammalian models compared to the human model (H. sapiens, Recon 1).
| Approach A | Approach B | |||||||
|---|---|---|---|---|---|---|---|---|
| Compartm. | Non-Compartm. | Compartm. | Non-Compartm. | |||||
| Organism | N | % | N | % | N | % | N | % |
| H. sapiens | 3189 | 2037 | 957 | 1280 | ||||
| P. troglodytes | 1566 | 49% | 1074 | 53% | 394 | 41% | 688 | 54% |
| R. norvegius | 2113 | 66% | 1503 | 74% | 726 | 76% | 911 | 71% |
| M. musculus | 2753 | 86% | 1720 | 84% | 851 | 89% | 1127 | 88% |
| B. taurus | 1716 | 54% | 1129 | 55% | 556 | 58% | 787 | 61% |
| C. lupus fam. | 1971 | 62% | 1249 | 61% | 627 | 66% | 842 | 66% |
Absolute number (N) and percentage (%) of active reactions (with at least one non-zero flux) for both compartmentalized and non-compartmentalized models created using approach A and B.
Identification of unique metabolic functions in the mouse
Given the high number of reactions mapped to the draft mouse reconstruction, and the high number of reactions with non-zero fluxes resulting in the corresponding metabolic modelA and modelB, we sought to finalize the mouse reconstruction by gap analysis/filling. First, we aimed to understand the metabolic differences between human and mouse to ensure that the mouse reconstruction is not merely a modified version of human metabolism. This implies that also genes and pathways unique to mouse metabolism needed to be identified. Therefore, we employed the Comparative Pathway Analyzer 1.0 [30] relying on the KEGG database [31] to extract metabolic maps displaying the existence of enzymes in both mouse and human for 66 out of 99 Recon 1 subsystems (Additional file 2). We then used these maps to perform a manual search of potential gaps between human and mouse metabolism. Out of 1550 reactions present within these subsystems, 1492 (96%) reactions existed in both species, 46 (3%) reactions existed only in humans and 1 (1%) reactions existed only in the mouse (Additional file 2). For the 25 remaining Recon 1 subsystems (excluding transporters), we did an extensive literature search for differences between the two species. Of those, only cytochrome P 450 metabolism was reported to differ significantly between mouse and human [32]. These defined differences will guide the subsequent gap filling process. However, since most of the metabolic functions seemed to be present in both human and mouse, we can employ the validation tests which were designed to evaluate the predictive potential of Recon 1 [17].
Gap filling of the mouse metabolic reconstruction
First, we decided to test which of the four mouse metabolic models were able to produce biomass by optimizing for the corresponding reaction accounting for all known biomass precursors required for cell replication. The compartmentalized and non-compartmentalized draft mouse models from approach A produced biomass without any modifications. In contrast, both versions of modelB were unable to produce biomass. Therefore, we sought to gap fill the draft non-compartmentalized modelB to identify reactions that needed to be added to the model for production of all biomass components. The SMILEY gap filling algorithm identifies reactions from a database that need to be added to a model to fulfill the optimality condition (e.g. production of biomass) [7].. Using the SMILEY algorithm, we searched for the minimal number of reactions within the entire KEGG database necessary to add to the non-compartmentalized mouse modelB in order to enable the production of biomass.
The results from 40 iterations of the algorithm were checked manually for two criteria: i) the result did not suggest a reversible reaction for a known irreversible reaction and ii) the added reaction(s) are known to occur in mouse. Of the 40 iterations, 30 suggested adding the same reaction either alone or in addition to some other metabolic reactions. This reaction (KEGG ID R06522) exists in mouse and humans (according to KEGG and Entrez gene [33]) but was not included in Recon1. It is a phosphohydrolase reaction involved in sphingolipid metabolism. Adding this reaction resulted in the ability of the refined non-compartmentalized modelB model to produce biomass at a similar rate as the non-compartmentalized modelA. The addition did, however, not result in biomass production in the compartmentalized modelB. Subsequently, we decided to focus the remainder of the study on modelA, as it captures most of the known metabolic capabilities in the mouse. ModelB was not further developed since it is missing a significant fraction of metabolic reactions and therefore does not function.
Validation of the mouse metabolic reconstruction
To validate the mouse reconstruction and to ensure mouse-characteristic metabolic properties of the resulting models, we manually tested each of the 288 FBA validation tests that were developed for Recon 1 [17] using both versions of modelA. Our literature survey on mouse metabolism, also focusing on the mouse essentiality of the 288 human tests, revealed no tests that were essential only in the human metabolism. Furthermore we found no evidence of mouse-specific additional tests of essentiality that could be added to our validation process. Therefore, we evaluated manually if all required reactions for each test are present in the mouse metabolism using Comparative Pathway Analyzer 1.0. A total of 260 tests passed the requirements and were therefore used for validation of the compartmentalized mouse modelA (Additional file 3). For the non-compartmentalized version of modelA, six additional tests were removed since they were compartment specific, resulting in 254 validation tests.
We ran the validation tests on the compartmentalized mouse draft modelA in an iterative manner and evaluated particularly the failed tests. This process revealed that out of the 131 reactions initially removed due to missing homologous genes in the HomoloGene database, 36 had sequence and physiological evidence of existence in the mouse (according to KEGG and EntrezGene databases) (Additional file 4). Those 36 reactions were therefore added again to the model, but the remaining 95 reactions were left out of the final model due to missing physiologic or sequence evidence. This addition resulted in a finalized mouse modelA that passed all the 260 validation tests. Also, we added the reaction discovered by the gap filling of modelB (KEGG ID R06522). Furthermore, unique mouse reactions which do not lead to metabolic dead ends in the model were added (KEGG IDs R03184, R00647 and R01465). The non-compartmentalized modelA similarly passed 100% of its 254 validation tests after the addition of these reactions. We also determined the functionality of modelB, even though the compartmentalized version cannot produce biomass. The compartmentalized and non-compartmentalized models created via approach B passed 50% and 85% of the validation tests respectively.
Properties of iMM1415
The resulting compartmentalized mouse metabolic reconstruction (Additional file 5), created by curation of modelA, termed iMM1415, contains a total of 3,724 reactions and 1,415 metabolic genes (Additional file 6 contains detailed description of all reactions in iMM1415, including the biomass reaction). Analysis of the reactions removed from Recon 1 during the creation of iMM1415 revealed that the greatest absolute number of non-included reactions in the mouse reconstruction were within steroid metabolism (16 out of 46 reactions) and blood group biosynthesis (14 out of 46 reactions). However, the largest percentage of non-included reactions was within the subsystems of stilbene, coumarine and lignin biosynthesis (50%, 1 out of 2 reactions) and limonene and pinene degradation (50%, 3 out of 6 reactions). This data suggests that simulation results from these metabolism subsystems will be more likely to be inaccurate and points out areas for future improvement of the mouse model. In the remainder of the paper, we will use models derived from iMM1415 for computations.
Comparison of iMM1415 with published mouse metabolic networks
Table 2 provides a comparison between our reconstruction and other published reconstructions, the most detailed to date being the recently published mouse metabolic network by Selvarasu et al [24]. Our model functions in eight cellular compartments compared to three compartments in most reconstructions. It has the most detailed description of mitochondria metabolism to date but fewer reactions in the cytosol. Furthermore iMM1415 contains the first attempt to describe metabolism in the golgi apparatus, lysosome, ribosome, peroxisome and nucleus. Due to the compartmentalization, a much greater number of transport reactions was required in our reconstruction compared to the previous ones. As less biochemical data exists on transport reactions a larger number of non-gene associated reactions are needed in our model than in the other models. Albeit difficult to compare directly due to different definition of minimum growth medium and biomass, our model predicts fewer essential genes but a higher percentage of them have been experimentally verified than in the other models published to date (Table 2).
Table 2.
Properties of iMM1415 and comparison with existing models
| iMM1415 | Selvarasu et al (2010). | Evsikov et al (2009). | Quek et al (2008). | Sheikh et al (2005). | |
|---|---|---|---|---|---|
| Dominant reconstruction method | Manual | Manual | Automatic | Automatic | Manual |
| Genes | 1,415 | 724 | 1,060 | 1,399 | 473 |
| Reactions | 3,724 | 715 | 2,018 | 1,757 | 0 |
| Compounds | 2,774 | 1,285 | 1,377 | 2,104 | 872 |
| Compartments | 8 | 3 | 1 | 3 | 3 |
| Reactions | 3,726 | 1,494 | 2,018 | 2,037 | 1,220 |
| Demand& Exchange& Biomass reactions | 455 | 0 | 0 | 0 | 0 |
| Cytosol | 931 | 1,085 | NA | 1,650 | 618 |
| Mitochondrial | 409 | 161 | NA | 387 | 12 |
| Extracellular | 473 | NA | NA | NA | NA |
| Golgi | 250 | NA | NA | NA | NA |
| Lysosome | 194 | NA | NA | NA | NA |
| Ribosome | 184 | NA | NA | NA | NA |
| Peroxisome | 95 | NA | NA | NA | NA |
| Nucleus | 88 | NA | NA | NA | NA |
| Transport | 1,101 | 248 | 0 | 64 | 267 |
| Non-gene associated reactions | 1,514 | 291 | 0 | 148 | 324 |
| Essential genes according to modela | 53a | 109a | NA | NA | 72 |
| Of which experimental data available | 17 | 20 | NA | NA | NA |
| % match to experimental data | 82% | 70% | NA | NA | NA |
| Produces biomass | 1 | 1 | 0 | 1 | 1 |
aunder "minimum growth" medium, differences exist in both biomass and minimum growth medium definitions between publications.
NA - not available
Essentiality of mouse metabolic genes
Given the high degree of functionality of iMM1415 we decided to employ it for phenotype simulations and to compare the in silico results to published experimental results from knockout mice or mice with mutations in metabolic genes. First, we performed a simulation of single gene knockouts for all genes in iMM1415 in order to determine in silico gene essentiality. A minimal growth medium supplemented with glucose was used for this simulation (as defined in additional file 7). A total of 53 genes were found to be essential as their deletion resulted in zero biomass production (Additional file 8). We found information on homozygous knockout phenotype for 17 of those genes in literature (Table 3). Of those, 14 (82%) genes had a confirmed lethal phenotype, whereas the remaining three (18%) had non-lethal phenotypes (Table 3). The majority of the genes with a predicted and confirmed essentiality status were within the cholesterol metabolism. This result indicates that cholesterol metabolism is especially vulnerable to mutations and environmental insults, perhaps due to the inability of the cholesterol metabolism to overcome disruption in metabolic reactions via alternative pathways.
Table 3.
Results on gene essentiality predictions by the finalized mouse model.
| Gene Name | Reactions | Subgroup | Mutation lethal | Comment | Reference |
|---|---|---|---|---|---|
| EBP | 3-beta-hydroxysteroid-delta(8),delta(7)-isomerase | Cholesterol Metabolism | yes | X-linked genes, homozygous males are non-viable (prenatal lethality) | Means et al. |
| DHCR7 | 7-dehydrocholesterol reductase | Cholesterol Metabolism | yes | Prenatal lethality of homozygotes | Yu et al. |
| DHCR24 | 24-dehydrocholesterol reductase | Cholesterol Metabolism | yes | Prenatal lethality of homozygotes | Mirza et al. |
| FDFT1 | Squalene synthase | Cholesterol Metabolism | yes | Prenatal lethality of homozygotes | Tozawa et al. |
| HSD17B4 | C-3 sterol keto reductase, Beta oxidation of long chain fatty acid, 3-hydroxyacyl-CoA dehydrogenase, hydroxysteroid (17-beta) dehydrogenase 4, peroxisomal lumped long chain fatty acid oxidation | Cholesterol Metabolism | yes | Pre/Peri/Postnathal lethality of homozygotes | Huyghe et al. |
| NSDHL | C-3 sterol dehydrogenase, C-4 methyl sterol oxidase | Cholesterol Metabolism | yes | X-linked gene, males and homozygous females are non-viable (prenatal lethality) | Cunningham et al |
| SC5DL | Lathosterol oxidase | Cholesterol Metabolism | yes | Perinatal lethality of homozygotes | Krakowiak et al. |
| SPTLC1 | serine palmitoyltransferase, long chain base subunit 1 | Sphingolipid Metabolism | yes | Embryonic lethality of homozygotes | Hojjati et al. |
| DHFR | dihydrofolate reductase, folate reductase | Folate Metabolism | yes | Embryonic lethality of homozygotes | Di Pietro et al. |
| PISD | phosphatidylserine decarboxylase, mitochondrial | Glycerophospholipid Metabolism | yes | Embryonic lethality of homozygotes | Steenbergen et al. |
| PHGDH | phosphoglycerate dehydrogenase | Glycine, Serine, and Threonine Metabolism | yes | Embryonic lethality of homozygotes | Yoshida et al. |
| HMGCR | Hydroxymethylglutaryl CoA reductase (ir) | Cholesterol Metabolism | yes | Embryonic lethality of homozygotes | Tanaka et al. |
| CBS | cystathionine beta-synthase, selanocystathionine beta-synthase | Methionine Metabolism | yes | Homozygous mice die within 5 weeks after birth | Watanabe et al. |
| SPTLC2 | serine C-palmitoyltransferase | Sphingolipid Metabolism | yes | Embryonic lethality of homozygotes | Hojjati et al. |
| PAH | L-Phenylalanine,tetrahydrobiopterin:oxygen oxidoreductase | Tyr, Phe, Trp Biosynthesis | no | Homozygous mice with dysruptions in the gene are viable | http://www.informatics.jax.org |
| TM7SF2 | C-14 sterol reductase | Cholesterol Metabolism | no | Although a mixture of mutations can be lethal, homozygotes for mutations are viable | http://www.informatics.jax.org |
| Gpam | glycerol-3-phosphate acyltransferase | Triacylglycerol Synthesis | no | Homozygous mice are viable | Howerton et al. |
Second, we searched the Mouse Genome Informatics database [27] for mouse reconstruction genes where a phenotype had been described with the word "lethality". Furthermore, we required that these genes were homozygous for a gene knockout and had either an embryonic or prenatal lethal phenotype. A total of 88 genes were identified this way. Five genes overlapped with the in silico essential genes (FDFT1, SPTLC1, PHGDH, HMGCR, CBS, SPTLC2). The observed discrepancy in lethality can be explained by either, i) growth environment simulations, ii) incomplete biomass reaction, iii) missing regulation, iv) wrongly included reactions, or v) any combination of the aforementioned possibilities. Wrongly included reactions could be identified by systematically eliminating non-gene-associated reactions from iMM1415 (e.g., using GrowMatch algorithm [34]) to improve the prediction of lethality. In contrast, the two genes that were essential in silico but in vivo non-essential for growth suggest missing functions in the metabolic network. Using SMILEY, or related algorithms, it might be possible to identify missing candidate genes.
Prediction of normal phenotypes
To further evaluate the predictive potential of iMM1415, we identified genes from the Mouse Genome Informatics database [27], for which a null mutation resulted in a normal phenotype. A total of eight such genes were found that were also present in the mouse reconstruction (OCRL, ACO1, PAFAH1B3, PGM1, FUT9, RHBG, ITPKC, SORD). For five of those, a gene deletion had no effect in the model, since isozymes existed within the reconstruction. For the three remaining genes (PGM1, FUT9, SORD), the deletion led to elimination of corresponding reactions. The effect of these deletions on growth depends on i) the presence of alternative reactions/pathways or ii) importance of reactions for biomass precursor synthesis. We investigated the effect of these three gene deletions on the overall network by performing a FVA. We then compared the FVA results of essential gene knockout (DHCR7) to the FVA results of wild type (Table 4) by considering only major sections of metabolism (Table 4). The largest perturbation resulted from the knockout simulation of the essential gene DHCR7, where seven major subsections of metabolism changed significantly. The knockout simulation of PGM1 led to significant changes within six major subsections of metabolism. The knockout simulation of the FUT9 gene led to significant changes within two major subsections of metabolism and the knockout simulation of SORD led to no significant changes within the major subsections of metabolism. Therefore, we suggest that six out of the eight genes are not likely to have noticeable metabolic phenotypes under our minimal medium condition due to presence of isozymes or alternative reactions/pathways.
Table 4.
Results from Flux Variability Analysis (FVA) of 3 knockout models for genes with a confirmed normal phenotype (PGM1, FUT9, SORD) and one essential gene (DHCR7).
| PGM1 | FUT9 | SORD | DHCR7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| down | up | p-value | down | up | p-value | down | up | p-value | down | up | p-value | |
| Amino Acid Metabolism | 61 | 160 | <0.00001 | 85 | 133 | 0.00115 | 122 | 98 | 0.10560 | 68 | 152 | <0.00001 |
| Carbohydrate Metabolism | 35 | 55 | 0.03501 | 48 | 48 | 1.00000 | 40 | 55 | 0.12380 | 22 | 75 | <0.00001 |
| Cofactor and Vitamin Metabolism | 20 | 75 | <0.00001 | 53 | 45 | 0.41900 | 60 | 37 | 0.01953 | 25 | 71 | <0.00001 |
| Energy Metabolism | 12 | 34 | 0.001180 | 19 | 27 | 0.23820 | 27 | 19 | 0.23820 | 13 | 32 | 0.00462 |
| Glycan Metabolism | 60 | 283 | <0.00001 | 118 | 229 | <0.00001 | 166 | 182 | 0.39110 | 49 | 294 | <0.00001 |
| Lipid Metabolism | 109 | 243 | <0.00001 | 165 | 189 | 0.20210 | 196 | 162 | 0.07234 | 128 | 225 | <0.00001 |
| Nucleotide Metabolism | 64 | 103 | 0.00255 | 80 | 91 | 0.40020 | 87 | 90 | 0.82160 | 45 | 122 | <0.00001 |
| Other Amino Acids Metabolism | 6 | 19 | 0.00932 | 11 | 14 | 0.54850 | 12 | 13 | 0.84150 | 7 | 18 | 0.02781 |
| Secondary Metabolites Metabolism | 0 | 0 | 1.00000 | 0 | 0 | 1.00000 | 0 | 0 | 1.00000 | 0 | 0 | 1.00000 |
| Transporters | 368 | 437 | 0.01502 | 397 | 422 | 0.38240 | 393 | 443 | 0.08376 | 337 | 479 | <0.00001 |
For each reaction within the model, FVA determines the minimum and maximum flux resulting in the optimal solution for the objective function (here: biomass production). Only those solutions that resulted in the same maximum biomass yield as the wild type model were used for the analysis. By comparing the flux range (the span between vmax,i and vmin,i for each reaction i) between the knockout model and a wild type model, each reaction was assigned a status of decreased (down), non-changing or increased (up) flux capacity (representing metabolic activity). For each metabolism subsystem, the number of reactions with decreased and increased flux capacity are shown. Significance testing was done by performing a single value Chi-Square test with 1 degree of freedom comparing against even probability of increased and decreased flux capacity. Significant p-values (p < 0.05/10) are typeset in bold.
Analysis of lipoprotein lipase deficiency on in silico phenotype
Finally, we sought to simulate knockouts of genes with softer phenotypes. The LPL gene encodes lipoprotein lipase (EC 3.1.1.34) [35], an enzyme that hydrolyzes chylomicrons and very low-density lipoproteins (VLDL) into free fatty acids. Individuals born with lipoprotein lipase deficiency have elevated levels of triglycerides and VLDL and suffer from recurrent episodes of abdominal pain and pancreatitis as well as eruptive xanthomas of the skin [35]. Mutations have been associated with increased risk of ischemic heart disease in man [36]. Mice without lipoprotein lipase are born with greatly elevated levels of triglycerides and VLDL, and after nursing, triglyceride levels soon become extremely high. Heterozygotes for the null mutation of LPL survive until adulthood but with elevated triglyceride levels [37].
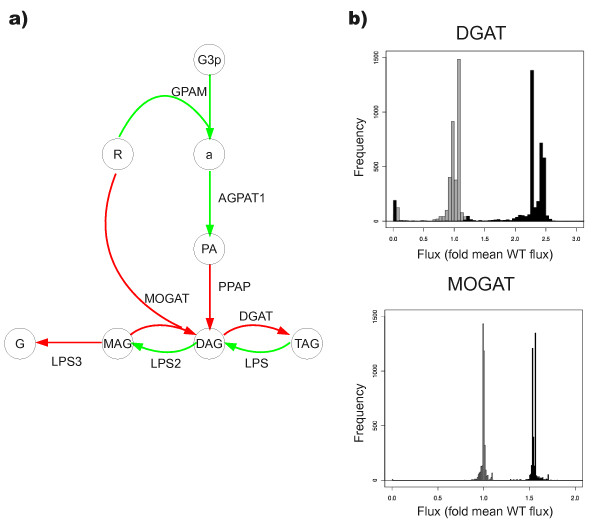
We simulated the knockout of the LPL gene and performed FVA to determine the effect of the gene deletion to the network properties. The largest perturbation was observed within i) the glycan metabolism, where 31 reactions had decreased and 312 reactions increased flux capacity (p < 10-16 against even probability of decreased and increased flux capacity); and ii) lipid metabolism, where 100 reactions with decreased and 253 reactions with increased flux capacity (p < 10-15 against even probability of decreased and increased flux capacity). Additionally, the flux capacity changed significantly within the triacylglycerol metabolism (Figure 2). The results suggest that there is increased flux capacity towards triglyceride synthesis while there is decreased flux capacity towards triglyceride degradation. The simulation results are therefore consistent with hypertriglyceridemia resulting from LPL mutations, as previously demonstrated with the mouse knockout model [37-51].
Figure 2.
Results from comparison of the flux variability analysis (FVA) for a LPL knockout model with a wild type model. Results from comparison of the flux variability analysis (FVA) for a LPL knockout model with a wild type model. a) A part of triacylglycerol metabolism is shown. Reactions with increased flux capacity in the knockout model are shown in red and reactions with decreased flux capacity are shown in green. b) The distribution of fluxes in FVA of a wild type model (dark gray) compared to knockout model (black) indicates increased flux through the diacylglycerol acetyltransferase (DGAT) and monoacylglycerol acetyltransferase (MOGAT). These results suggest that hypertriglyceridemia can result from knockout of the LPL gene. Reactions: AGPAT1 - 1-acylglycerol-3-phosphate O-acyltransferase 1; DGAT - diacylglycerol acyltransferase; GPAM - glycerol-3-phosphate acyltransferase; LPS - lipoprotein lipase LPS2 - lipoprotein lipase 2; LPS3 - lipase; MOGAT - monoacylglycerol acyltransferase; PPAP - phosphatidic acid phosphatase. Metabolites: a-lysophosphatidic acid; DAG - diacylglycerol (Homo sapiens); G - glycerol; G3p - Glycerol 3-phosphate; MAG - monoacylglycerol 2; PA - phosphatidic acid; R - R groups (total); TAG - triacylglycerol.
Discussion
Can the success of metabolic systems biology in microbes be repeated for more complex organisms? A prerequisite for such success is the existence of high quality metabolic network reconstructions. Here, we examined if the detailed and validated network reconstruction of human metabolism, Recon 1, could be used as a basis for reconstructing metabolic networks in other mammals. We then focused on mouse to create a manual curated, comprehensive reconstruction of mouse metabolism that was subsequently employed to determine normal and lethal growth phenotypes. Overall, we found good agreement between the model's prediction and reported mouse phenotypes, suggesting that the mouse reconstruction has similar quality properties as Recon 1.
Initial analysis of sequenced mammalian genomes revealed that the highest number of genes orthologous to Recon 1 genes was found in the mouse while the lowest number was present in the chimpanzee (Table 1). This result was surprising since the sequence homology between human and chimpanzee is ~99% [29] while the sequence homology between human and mouse is in the range of 85-92% [52,53]. This observation reflects the fact that many enzymes involved in human metabolism were discovered by analyzing their mouse orthologues, leading to a high likelihood that mouse metabolic enzymes are known in human and thus present in Recon 1.
The initial search for homologous genes indicated that a functional mouse reconstruction could be produced based on Recon 1. Using two different approaches, we created four versions of a draft mouse model (Figure 1). These draft models were then completed by automated and manual gap filling analysis and validated against 260 FBA-based tests. This effort resulted in iMM1415, which accounts for more genes and reactions, distributed over eight cellular compartments, than previously published reconstructions (Table 2) in addition to its representation of eight cellular compartments. Thus, the mouse metabolic reconstruction presented herein is the most comprehensive reconstruction available to date. However, its usage of the human Recon 1 for reactions information (including cellular location and reversibility) risks that the reactions included are skewed towards human metabolism and that reactions unique to the mouse might be missed.
A surprisingly high number of non-gene-associated reactions are included in Recon 1. Many of those are intra-cellular transport reactions, demand or exchange reactions with sparse literature supporting their existence. They were included to provide a functional model of human metabolism across the eight cellular compartments and were therefore also added to iMM1415. We feel that the gain for a highly functional model by their full inclusion supersedes the risk of artificially inflating the properties of the mouse reconstruction. Furthermore, 30-40% of all known enzymes with an EC number are orphan meaning that their biochemical activity is well characterized while the encoding gene is unknown in any organism [54]. Many of these enzymes (13-37%) are within major metabolism pathways [55] suggesting that metabolic reconstructions of even well-annotated organisms will have a set of non-gene associated reactions in order to ensure their correct functionality. This observation also argues against reconstructing or analyzing metabolic networks based solely on genomic data as they are likely to have numerous missing metabolic functions. Also, the non-gene associated reactions are an extremely interesting area of future research as they suggest pathways and enzymes that require further experimental exploration [55]. Furthermore, several reactions leading to metabolic dead ends in the model were included in Recon 1 and subsequently in our mouse reconstruction. They highlight missing links in knowledge and form testable hypotheses that should be further investigated. Any effort of a consensus metabolic reconstruction of the mouse should focus on reviewing the non-gene-associated reactions and those leading to dead-ends in the model in detail.
Our approach could in theory also be used to construct high quality metabolic reconstructions for other mammals. Given our results (Table 1), it is likely that a rat reconstruction could be obtained using a similar approach as presented here (Figure 1). However, the resulting network is expected to be less complete, limiting its application and requiring more manual curation. Metabolic reconstructions of other mammals will be of insufficient quality and predictive potential using the presented approach due to the low number of homologous genes with Recon1 genes.
The phenotype prediction properties of metabolic models have been extensively applied for microorganisms, however, only limited studies exists for mammals [20,56,57]. The mouse is an ideal model organism for testing and validating phenotypic properties since multiple inbred strains, several thousand gene knockout strains and various cell lines exist [27] including knockouts of many metabolic genes. We found that the majority of predicted essential genes also had a lethal phenotype in vivo. Interestingly, the majority of the in silico essential genes were found to be within cholesterol metabolism, indicating both the importance and mutation vulnerability of this metabolism pathway. This observation is in concordance with results based on an earlier mouse metabolic model, where the majority of essential genes were from cholesterol metabolism [24]. These results should guide further research and might be helpful in understanding human disease based on lipid abnormalities, such as atherosclerosis [24]. However, many more genes were identified in mouse knockout databases [27] which result in a lethal phenotype in vivo but had a non-lethal phenotype in silico. The disagreement could result from the non-tissue specificity of iMM1415 or incomplete biomass reaction and highlights starting points for future research to further our insight into mouse metabolism.
The majority of genes for which an in vivo experimental knockout strain has been developed have a non-lethal phenotype. Similarly, in silico knockout simulations resulted in little or no perturbation of the mouse metabolic network. Recently, it was shown for yeast that even though deletion of ~80% of yeast genes resulted in no apparent phenotype when grown in rich medium, a measurable growth phenotype was observed for ~97% of yeast genes when the medium was either depleted of certain ingredients or biologically active compounds added to the medium [58]. Furthermore, virtually all genes were essential under some growth conditions [58]. It is an important subject of future research to see if the mammalian models suggest similar findings, as such analysis might further understanding of both complex genotypes and the effects of environmental factors on disease pathogenesis.
Conclusions
Here, we have created a reconstruction of mouse metabolism based on sequence homology, using the highly detailed Recon 1 of human metabolism as a basis for our reconstruction. The model has been rigorously validated using 260 flux balance analysis based tests. The resulting reconstruction, iMM1415, is to date the most comprehensive reconstruction of mouse metabolism. Our phenotype simulation results suggest that the current quality of both the human Recon 1 and the derived iMM1415 models are sufficient for phenotype predictions. The existence of these two reconstructions should encourage the creation of detailed metabolic network reconstructions for other mammals. With the ongoing international knockout mouse project http://www.knockoutmouse.org aiming to produce knockout of all protein coding genes within the mouse genome and the Collaborative Cross project aiming to crossbreed eight inbred strains of mice [59], the validation of mammalian metabolic models should become extensive. Also, the sequencing of more inbred strains of experimental mice will provide new opportunities in studying the effects of genetic variability on metabolism, utilizing the mouse reconstruction as a data analysis platform. The existent and emerging reconstructions should be joined in a collaborative effort to reach a consensus reconstruction of the mouse metabolism to maximize its accuracy and utilization properties. Reconstruction jamboree meetings have been held for various organisms to obtain consensus metabolic reconstructions and to increase their content in a community driven approach [4,60]. Thoroughly validated genome-scale reconstructions should provide a broad platform for studying mammalian metabolism and can form the basis for developing therapeutic interventions related to pathological metabolic states [38,42-44,48,51,61].
Methods
All model calculations were done in MATLAB version 2009a (The Mathworks Inc.) using the COBRA toolbox [6] and the Mosek linear solver (Mosek ApS, Denmark). Statistical tests and figure preparation were done in R, version 2.9.1 (The R foundation, Austria). In-house scripts for data processing were written in Java and are available upon request from the authors.
Flux balance analysis and flux variability analysis
Given a reconstructed metabolic network on a mathematical form with appropriate constraints on each reaction (representing reversibility, maximum and minimum flux etc.) and a biological objective Z (such as biomass), flux balance analysis (FBA) calculates a set of network fluxes that maximize Z. It has several derived applications used to further analyze the flow of metabolites through a metabolic model [62]. One such application is flux variability analysis (FVA) [63]. FVA uses linear programming methods to find the minimal and maximal flux values that can be achieved by each reaction while contributing to maximal production of the objective reaction.
Creation of draft reconstructions
The entire HomoloGene build 52 was downloaded on 11/17/2008 [28]. We extracted data from the dataset for all 1,496 genes in Recon 1 [17]. For each of the 20 species in the HomoloGene database, we then checked for the availability of a homologous gene for each Recon 1 gene. Next we created draft reconstructions for each species using two different approaches as described in the result section. The draft reconstructions were converted into condition-specific draft models [4]. All draft models were checked for functionality by optimizing for biomass while allowing unrestricted uptake of metabolites (vmin, metabolite ≥ -1000 mmol/gdw hr). FVA was performed on the draft models to determine number of reactions with a non-zero flux. Following FVA on each draft model, we drew phylogenetic trees of the results and compared to phylogenetic tree based on all transcripts of protein domain sequence from the SuperFamily database [64].
Gap filling
Gap filling was done using the SMILEY algorithm, as described elsewhere [7]. We downloaded and used the entire KEGG database (accessed at 9/15/2009) [31] as universal reaction database. Additionally, we created a database of transport and exchange reaction for all metabolites in Recon1 and the KEGG database. These two databases were used to determine candidate reactions to fill gaps in the non-compartmentalized draft model. The gap filling algorithm was set to run 40 iterations. Each solution was then manually checked for applicability (such as the feasibility of the suggested directionality) before choosing a proposed solution.
Validation
For validation of the mouse reconstruction, we used a modification of the validation process used in the creation of the human Recon 1 network. Each of the 288 validation tests used for Recon 1 was manually checked for applicability in the mouse. This was done by manually reviewing each validation test using Comparative Pathway Analyzer 1.0 [30]. If all enzymes participating in validation test of choice existed both in human and mouse, the test was used in the mouse validation. This resulted in 260 validation tests for the mouse model. The model was then improved manually in an iterative manner by checking the reactions involved in failed tests until it passed all validation tests.
Simulations
Information on mouse knockout phenotypes were gained from the Mouse Genome Informatics site (http://www.informatics.jax.org, accessed on 1/12/2010) [27]. For each gene whose knockout was simulated, a model with the corresponding reaction bounds set to zero was created. Both the wild type and knockout simulation type models allowed cellular uptake of vital amino acids, vital fatty acids, glucose, oxygen, hydrogen, sulfur oxide, phosphate and ions while optimizing for biomass (detailed exchange constraints listing is in Additional file 7). As computational methods do not allow direct sampling of the flux solution space for large networks[65,66], we performed FVA[63] to estimate the range of flux values for each reaction resulting in the optimal solution. By comparing the FVA for the wild type model and knockout models, each reaction was then assigned a status of decreased flux capacity (i.e., reduced vmax,i), no change or increased flux capacity. For an unbiased selection of interesting metabolic subsystems, we counted number of reactions with increased and decreased flux ranges and calculated a single value Chi-Square test with 1 degree of freedom comparing the observation against even probability of increased and decreased flux capacity. Since independent tests were done for each of the 10 major subsystems, a p-value of 0.05/10 was considered statistically significant to correct for multiple testing.
Authors' contributions
MIS, NJ, ES, IT and BOP conceived of the study. MIS, NJ and IT created and validated the models. MIS performed phenotype simulations. MIS drafted the manuscript. MIS, ES, IT and BOP revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental file S1: Absolute number and percentage of Recon 1 genes found in all species within the HomoloGene database.
Supplemental file S2: Details of KEGG search for unique reactions in mouse and human using Comparative Pathway Analyzer 1.0.
Supplemental file S3: List of 260 flux balance analysis tests used for validation of the mouse reconstruction.
Supplemental file S4: List of genes added again during the validation process and the corresponding metabolic subgroups.
Supplemental file S5: The iMM1415 model of mouse metabolism in a SMBL format.
Supplemental file S6: A detailed reaction list of all reactions in the iMM1415, including the biomass reaction.
Supplemental file S7: Uptake rates for exchange reactions in the mouse reconstruction under minimal medium conditions.
Supplemental file S8: A list of genes predicted to be essential in the mouse metabolic network, along with bibliographic information if available.
Contributor Information
Martin I Sigurdsson, Email: martiningi@gmail.com.
Neema Jamshidi, Email: neema@ucsd.edu.
Eirikur Steingrimsson, Email: eirikurs@hi.is.
Ines Thiele, Email: ithiele@hi.is.
Bernhard Ø Palsson, Email: bpalsson@bioeng.ucsd.edu.
Acknowledgements
This work was supported by ERC grant agreement No. 232816 - SYSTEM_Us between the European Research Council and Bernhard Ø. Palsson and a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme.
References
- Edwards JS, Palsson BO. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem. 1999;274:17410–17416. doi: 10.1074/jbc.274.25.17410. [DOI] [PubMed] [Google Scholar]
- Palsson B. Metabolic systems biology. FEBS Lett. 2009;583:3900–3904. doi: 10.1016/j.febslet.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Palsson BØ. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ND, Reed JL, Palsson BØ. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. Systems approach to refining genome annotation. Proc Natl Acad Sci USA. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Palsson BØ, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Thiele I, Weekes D, Li Z, Jaroszewski L, Ginalski K, Deacon AM, Wooley J, Lesley SA, Wilson IA, Palsson B, Osterman A, Godzik A. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science (80-) 2009;325:1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BØ, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T, Ma H, Zhao X, Goryanin I. Compartmentalization of the Edinburgh Human Metabolic Network. BMC Bioinformatics. 2010;11:393. doi: 10.1186/1471-2105-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Park J, Kay KA, Christakis NA, Oltvai ZN, Barabási A. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci USA. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgård MJ, Palsson BØ, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- Sigurdsson MI, Jamshidi N, Jonsson JJ, Palsson BO. Genome-scale network analysis of imprinted human metabolic genes. Epigenetics. 2009;4:43–46. doi: 10.4161/epi.4.1.7603. [DOI] [PubMed] [Google Scholar]
- Seo S, Lewin HA. Reconstruction of metabolic pathways for the cattle genome. BMC Syst Biol. 2009;3:33. doi: 10.1186/1752-0509-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L, Nielsen LK. On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform. 2008;21:89–100. full_text. [PubMed] [Google Scholar]
- Selvarasu S, Karimi IA, Ghim G, Lee D. Genome-scale modeling and in silico analysis of mouse cell metabolic network. Mol Biosyst. 2010;6:152–161. doi: 10.1039/b912865d. [DOI] [PubMed] [Google Scholar]
- Sheikh K, Förster J, Nielsen LK. Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Prog. 2005;21:112–121. doi: 10.1021/bp0498138. [DOI] [PubMed] [Google Scholar]
- Savinell JM, Palsson BO. Network analysis of intermediary metabolism using linear optimization. II. Interpretation of hybridoma cell metabolism. J Theor Biol. 1992;154:455–473. doi: 10.1016/S0022-5193(05)80162-6. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37:D712–9. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GAT, Gabriel SB, Reich D, Donnelly P, Altshuler D. Comparison of fine-scale recombination rates in humans and chimpanzees. Science (80-) 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Oehm S, Gilbert D, Tauch A, Stoye J, Goesmann A. Comparative Pathway Analyzer--a web server for comparative analysis, clustering and visualization of metabolic networks in multiple organisms. Nucleic Acids Res. 2008;36:W433–7. doi: 10.1093/nar/gkn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar VS, Maranas CD. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput Biol. 2009;5:e1000308. doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi M, Wilson DE, Iverius PH, Wu L, Hata A, Hegele R, Williams RR, Lalouel JM. Missense mutation (Gly----Glu188) of human lipoprotein lipase imparting functional deficiency. J Biol Chem. 1990;265:5910–5916. [PubMed] [Google Scholar]
- Wittrup HH, Tybjaerg-Hansen A, Steffensen R, Deeb SS, Brunzell JD, Jensen G, Nordestgaard BG. Mutations in the lipoprotein lipase gene associated with ischemic heart disease in men. The Copenhagen city heart study. Arterioscler Thromb Vasc Biol. 1999;19:1535–1540. doi: 10.1161/01.atv.19.6.1535. [DOI] [PubMed] [Google Scholar]
- Weinstock PH, Bisgaier CL, Aalto-Setälä K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg AD, Zechner R, Breslow JL. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham D, Spychala K, McLarren KW, Garza LA, Boerkoel CF, Herman GE. Developmental expression pattern of the cholesterogenic enzyme NSDHL and negative selection of NSDHL-deficient cells in the heterozygous Bpa(1H)/+ mouse. Mol Genet Metab. 2009;98:356–366. doi: 10.1016/j.ymgme.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Mol Cell Biol. 2002;22:4158–4166. doi: 10.1128/MCB.22.12.4158-4166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojjati MR, Li Z, Jiang X. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim Biophys Acta. 2005;1737:44–51. doi: 10.1016/j.bbalip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Howerton SB, Nagpal A, Williams LD. Surprising roles of electrostatic interactions in DNA-ligand complexes. Biopolymers. 2003;69:87–99. doi: 10.1002/bip.10319. [DOI] [PubMed] [Google Scholar]
- Huyghe S, Mannaerts GP, Baes M, Van Veldhoven PP. Peroxisomal multifunctional protein-2: the enzyme, the patients and the knockout mouse model. Biochim Biophys Acta. 2006;1761:973–994. doi: 10.1016/j.bbalip.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Krakowiak PA, Wassif CA, Kratz L, Cozma D, Kovárová M, Harris G, Grinberg A, Yang Y, Hunter AGW, Tsokos M, Kelley RI, Porter FD. Lathosterolosis: an inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. Hum Mol Genet. 2003;12:1631–1641. doi: 10.1093/hmg/ddg172. [DOI] [PubMed] [Google Scholar]
- Means GD, Boyd Y, Willis CR, Derry JM. Transgenic rescue of the tattered phenotype by using a BAC encoding Ebp. Mamm Genome. 2001;12:323–325. doi: 10.1007/s003350010262. [DOI] [PubMed] [Google Scholar]
- Mirza R, Hayasaka S, Takagishi Y, Kambe F, Ohmori S, Maki K, Yamamoto M, Murakami K, Kaji T, Zadworny D, Murata Y, Seo H. DHCR24 gene knockout mice demonstrate lethal dermopathy with differentiation and maturation defects in the epidermis. J Invest Dermatol. 2006;126:638–647. doi: 10.1038/sj.jid.5700111. [DOI] [PubMed] [Google Scholar]
- Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem. 2005;280:40032–40040. doi: 10.1074/jbc.M506510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Takeuchi T. Expression of tyrosinase gene in transgenic albino mice: the heritable patterned coat colors. Pigment Cell Res. 1992;5:300–303. doi: 10.1111/j.1600-0749.1992.tb00553.x. [DOI] [PubMed] [Google Scholar]
- Tozawa R, Ishibashi S, Osuga J, Yagyu H, Oka T, Chen Z, Ohashi K, Perrey S, Shionoiri F, Yahagi N, Harada K, Gotoda T, Yazaki Y, Yamada N. Embryonic lethality and defective neural tube closure in mice lacking squalene synthase. J Biol Chem. 1999;274:30843–30848. doi: 10.1074/jbc.274.43.30843. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Osada J, Aratani Y, Kluckman K, Reddick R, Malinow MR, Maeda N. Mice deficient in cystathionine beta-synthase: animal models for mild and severe homocyst(e)inemia. Proc Natl Acad Sci USA. 1995;92:1585–1589. doi: 10.1073/pnas.92.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Furuya S, Osuka S, Mitoma J, Shinoda Y, Watanabe M, Azuma N, Tanaka H, Hashikawa T, Itohara S, Hirabayashi Y. Targeted disruption of the mouse 3-phosphoglycerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J Biol Chem. 2004;279:3573–3577. doi: 10.1074/jbc.C300507200. [DOI] [PubMed] [Google Scholar]
- Yu H, Wessels A, Tint GS, Patel SB. Partial rescue of neonatal lethality of Dhcr7 null mice by a nestin promoter-driven DHCR7 transgene expression. Brain Res Dev Brain Res. 2005;156:46–60. doi: 10.1016/j.devbrainres.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Blatt C, DePamphilis ML. Striking homology between mouse and human transcription enhancer factor-1 (TEF-1) Nucleic Acids Res. 1993;21:747–748. doi: 10.1093/nar/21.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Swergold GD, Seldin MF. Examination of sequence homology between human chromosome 20 and the mouse genome: intense conservation of many genomic elements. Hum Genet. 2003;113:60–70. doi: 10.1007/s00439-003-0920-x. [DOI] [PubMed] [Google Scholar]
- Chen L, Vitkup D. Distribution of orphan metabolic activities. Trends Biotechnol. 2007;25:343–348. doi: 10.1016/j.tibtech.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Lespinet O, Labedan B. Orphan enzymes could be an unexplored reservoir of new drug targets. Drug Discov Today. 2006;11:300–305. doi: 10.1016/j.drudis.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BØ. Systems biology of SNPs. Mol Syst Biol. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol. 2009;5:263. doi: 10.1038/msb.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science (80-) 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-y. [DOI] [PubMed] [Google Scholar]
- Herrgård MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Blüthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novère N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K, Spasić I, Weichart D, Brent R, Broomhead DS, Westerhoff HV, Kirdar B, Penttilä M, Klipp E, Palsson BØ, Sauer U. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza R, Hayasaka S, Kambe F, Maki K, Kaji T, Murata Y, Seo H. Increased expression of aquaporin-3 in the epidermis of DHCR24 knockout mice. Br J Dermatol. 2008;158:679–684. doi: 10.1111/j.1365-2133.2007.08424.x. [DOI] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Palsson BØ. Use of randomized sampling for analysis of metabolic networks. J Biol Chem. 2009;284:5457–5461. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- Thiele I, Price ND, Vo TD, Palsson BØ. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280:11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental file S1: Absolute number and percentage of Recon 1 genes found in all species within the HomoloGene database.
Supplemental file S2: Details of KEGG search for unique reactions in mouse and human using Comparative Pathway Analyzer 1.0.
Supplemental file S3: List of 260 flux balance analysis tests used for validation of the mouse reconstruction.
Supplemental file S4: List of genes added again during the validation process and the corresponding metabolic subgroups.
Supplemental file S5: The iMM1415 model of mouse metabolism in a SMBL format.
Supplemental file S6: A detailed reaction list of all reactions in the iMM1415, including the biomass reaction.
Supplemental file S7: Uptake rates for exchange reactions in the mouse reconstruction under minimal medium conditions.
Supplemental file S8: A list of genes predicted to be essential in the mouse metabolic network, along with bibliographic information if available.