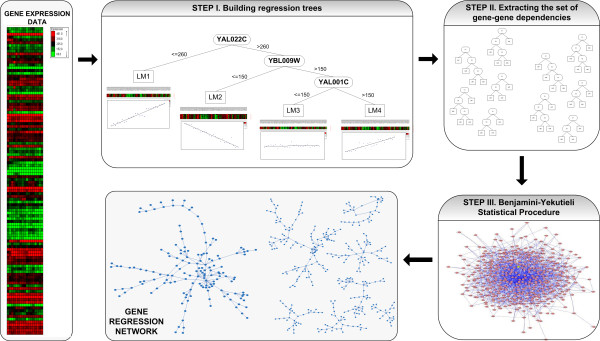
Figure 1.
Schematic view of the proposed method. In the first step, for each gene (target gene) a model tree is generated, which provides a partition of the space. Linear regression functions are built in the leaves of the tree. These linear regression functions can be seen as prototypes to estimate the value of the target gene. Genes involved in the linear regression functions might be identified as potential dependencies. This is an iterative process that is made for each gene taking the remaining genes as input to build the model tree, and it provides a set of hypothetical gene-gene interactions. Only the model trees with low prediction error will be conserved. Next, the Benjamini-Yekutieli statistical procedure is applied in order to assess the significance of the dependencies.