Fig. 2.
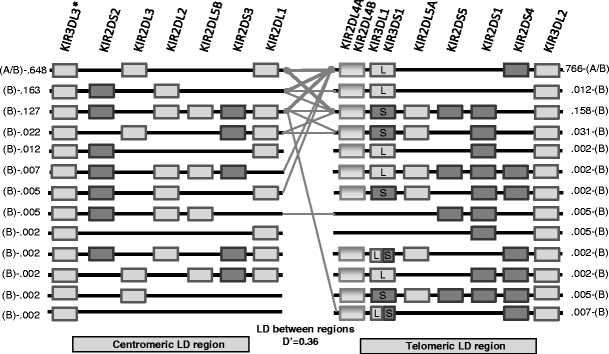
Compact illustration of the KIR genes framework haplotype using centromeric and telomeric regions. Lines indicate associations of telomeric and centromeric regions. Thin lines connect haplotypes whose frequency is between 0.3% and 10%. Thick lines connect haplotype whose frequency is greater than 10%. Rectangles indicate presence of the KIR gene, and lines represent absence of the KIR gene. All haplotype structures within the sample are displayed. This figure also presents a specific D′: the Hedrick’s multiallelic D′, which represents the degree of LD between two regions, treating each haplotype within a region as an allele. The display is adapted from the Haploview software haplotype blocks display. KIR3DL2 is always present at the telomeric end. (*) KIR3DL3 is assumed to be always present but was not typed at the allelic level in the families. All founders and offspring were positive for KIR3DL3. Because KIR3DL3 and KIR3DL2 appear to be always present, it is not possible to study LD for these genes in terms of their presence or absence. The physical gene position is a consensus map, and another order may better reflect the reality of a given haplotype. Although LD computations do not use the physical positions of the genes and their alleles, LD measures are not necessarily independent of gene order. For example, this is the case when one considers the possibility of 2DS3 and 2DS5 being present on either side or both sides of 2DL4. Frequencies and membership in the A or B haplotype are displayed on the side of the haplotypes. For example, in the centromeric region, the haplotype KIR3DL3–KIR2DL3–KIR2DL1 can belong to both A and B haplotypes and its frequency in our data is 0.648 (64.8%), it combines in more than 10% of the cases with KIR2DL4–KIR3DS1–KIR2DL5A–KIR2DS1–KIR3DL2 telomeric haplotype, which is found in B haplotypes and whose frequency is 15.8%
