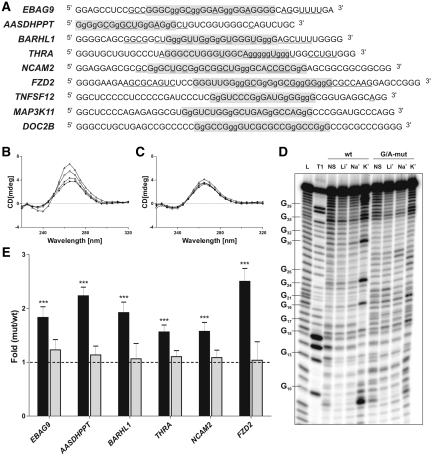
Figure 1.
Identification of G-quadruplex structures and translational repressors. (A) Sequences of the wild-type PG4s of the nine selected candidates. Sequences of the original potential G-quadruplexes are shaded in gray. The lowercase guanosines (g) correspond to those changed to adenosines in the G/A-mutants. Nucleotides that were cleaved significantly more in the presence of KCl, as compared to in the presence of LiCl, in the in-line probing experiments as determined by the quantification with the SAFA software, are underlined. (B, C) Circular dichroism spectra for the EBAG9 PG4 candidate using 4 µM of either the wild type (B) or the G/A-mutant (C) sequences and either in the absence of salt (close circles) or in the presence of 100 mM LiCl (closed triangles), NaCl (open circles) or KCl (open triangles). (D) Autoradiogram of a 10% denaturing (8M urea) polyacrylamide gel of the in-line probing of the 5′-labeled EBAG9 wild-type and G/A-mutant PG4 versions performed either in the absence of salt (NS), or in the presence of 100 mM LiCl, NaCl or KCl. The lanes designated L and T1 are an alkaline hydrolysis and a ribonuclease T1 (RNase T1) mapping of the wild-type version, respectively. Representative guanosine residues are indicated on the left of the gel. (E) Gene expression levels of the different constructs used at the protein level using either the luciferase assay (black bars) or the mRNA level as determined by RT-qPCR (gray bars). The x-axis identifies the candidates and the y-axis the fold difference that corresponds to the value obtained for the G/A-mutant version divided by that obtained for the wild type version for each candidate. The RT-qPCR values were obtained using the ΔΔCT method with the Fluc gene as internal control and the wild-type version as calibrator. Error bars were calculated using a minimum of three independent experiments. ***P < 0.001.