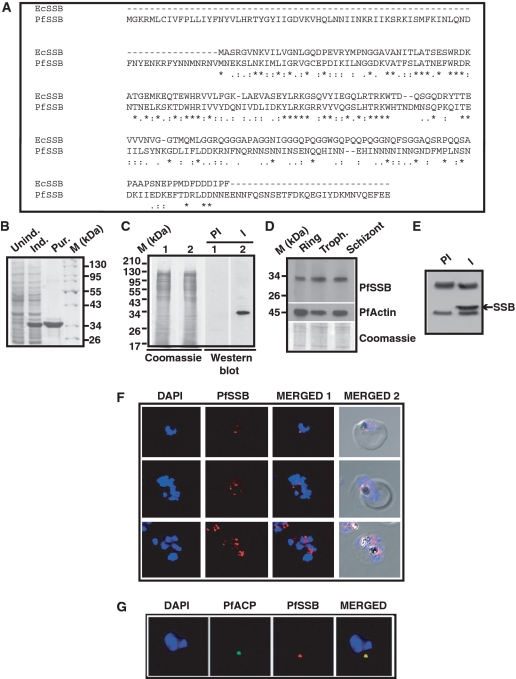
Figure 1.
Amino acid sequence alignment, protein expression profile and immunolocalization of PfSSB. (A) Clustal W amino acid sequence alignment of PfSSB with E. coli SSB (EcSSB). Identical residues are marked with ‘asterisk’ whereas ‘colon’ and ‘end note’ signs are used to indicate strongly and weekly similar residues, respectively. (B) Coomassie-stained gel picture of induction and purification of His6-tagged Pf SSB. Protein markers (M) are indicated on the right. (C) The right panel shows the western blot analysis using pre-immune (PI) or immune (I) sera against PfSSB (lanes 1 and 2) against 3D7 parasite lysate. The left panel shows the coomassie stained gel following transfer of the proteins on the membrane (loading control) (D) Expression profile of PfSSB at the different erythrocytic stages as indicated on top is shown by western blot analysis using polyclonal antibodies against PfSSB. The same blot was reprobed with antibodies against PfActin (middle panel). The bottom panel shows the coomassie stained gel following transfer. (E) Immunoprecipitation of PfSSB protein from parasite lysate using immune (I) or pre-immune sera (PI) followed by western blot analysis using polyclonal antibodies against SSB. Arrowhead indicates the position of PfSSB protein. (F) Localization of PfSSB by immunofluorosecnce analysis using glass slide containing fixed parasites obtained from trophozoite and schizont stages in the presence of anti-PfSSB antibodies. DAPI indicates nuclei whereas merged panel shows the presence of PfSSB with respect to the nuclei. The last panel shows the phase picture of parasite infected RBC. (G) Immuno-colocalization of PfSSB and apicoplast marker acyl carrier protein (ACP). Merged panel shows the co-localization of both the proteins.