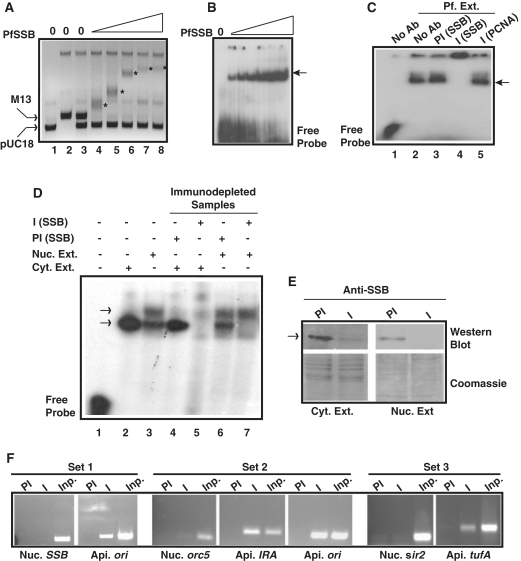
Figure 4.
In vitro and in vivo DNA binding property of Pf SSB. (A) PfSSB binds specifically to ssDNA. Different amount of Pf SSB was incubated with the mixture of M13 mp18 ssDNA and pUC18 dsDNA followed by separation of the protein-DNA complex by agarose gel electrophoresis. ssDNA was progressively retarded (asterisk) with increasing concentration of PfSSB whereas mobility of dsDNA was not affected. (B) Electrophoretic mobility shift assay using short radiolablled single-stranded oligonucleotide probe with increasing amount of Pf SSB. The arrowhead indicates the protein bound DNA. (C) Gel retardation analysis of single stranded DNA binding activity from P. falciparum cell extract. Labelled ssDNA probe was incubated with P. falciparum cell extract in the absence or presence of anti-PfSSB or anti-PCNA antibodies or pre-immune sera against SSB followed by PAGE analysis of the protein–DNA complexes (shown by arrowhead). Supershift of the DNA–protein complex was observed only in the presence of immune sera against PfSSB (lane 4) but not in the presence of pre-immune sera (lane 3) or non-immune sera (anti-PCNA) (lane 5). (D) Single-stranded DNA binding activity of cytoplasmic or nuclear extract or immunodepleted samples from each fraction in the presence of preimmune and immune sera against PfSSB. A single shifted band of labelled probe was observed in cytoplasmic fraction (lane 2) whereas two such bands were observed in nuclear extract treated samples (lane 3). No band was observed in the presence of immunodepleted cytoplasmic extract (lane 5) whereas the bottom band was not visible in the presence of immunodepleted nuclear extract treated sample (lane 7). Pre-immune depleted samples did not affect the pattern of bands (lanes 4 and 6) found in extract only lanes (lanes 2 and 3). (E) The upper part of the left panel shows the western blot analysis of immune and pre-immune depleted Plasmodium cytoplasmic extract whereas the lower part represents the coomassie stained gel as loading control following transfer of the proteins on the membrane. Similarly, the upper part of the right panel shows the western blot analysis of the immunodepleted nuclear extract (immune and pre-immune treated) and the lower part of the same panel represents the coomassie stained gel as loading control. The arrowhead indicates the position of the PfSSB protein. (F) Specific binding of PfSSB to apicoplast DNA in ChIP assay. PCR amplification of ori region of apicoplast (api.) DNA was found only in the immunoprecipitated samples using immune sera (I) against PfSSB but not with the pre-immune (PI) sera whereas nuclear (nuc.) ssb gene could not be amplified using ssb gene specific primers (set 1). Further, PCR amplification of immunoprecipitated DNA along with input (inp.) genomic DNA were performed using specific primer sets from nuclear orc5, apicoplast ori and IRA region (set 2) or nuclear sir2 and apicoplast tufA regions (set 3). Specific products were found only in the immunoprecipitated samples using immune sera and primer sets from apicoplast DNA. No significant amplification was found using primer sets from nuclear DNA. PCR products were obtained in all the control input lanes.