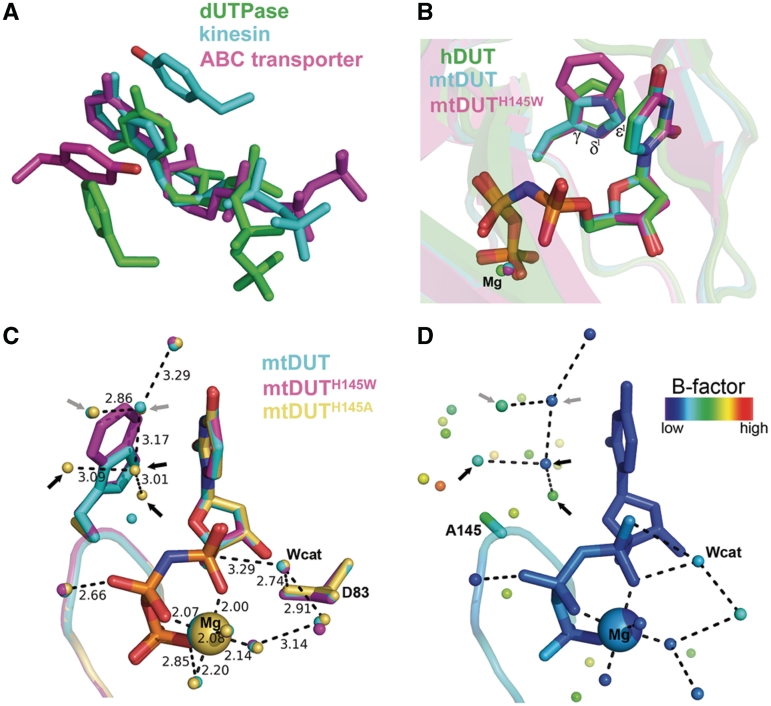
Figure 1.
Structural aspects of the enzyme–substrate π−π interaction in dUTPase and in other nucleotide hydrolases. (A) The π−π interactions between enzyme and substrate in representatives of various nucleotide hydrolase families [PDB IDs 2HQU (13), 2NCD (29) and 1XEF (30)]. (B) Structural superimposition of dUTPase active sites conferring different aromatic amino acids [Phe, His, Trp in PDB IDs 2HQU (13), 2PY4 (16), 3HZA, respectively]. (C) Superimposition of the newly acquired mutant structures with wild-type mtDUT. Grey and black arrows point to water molecules found in both mtDUT and mtDUTH145A but not in mtDUTH145W or only in mtDUTH145A, respectively. D83 is the catalytic residue that polarizes the nucleophile attacking water molecule (Wcat) (25). (D) Same view of the mtDUTH145A active site colored by B factors. Note the relatively low mobility of the Trp-replacing waters.