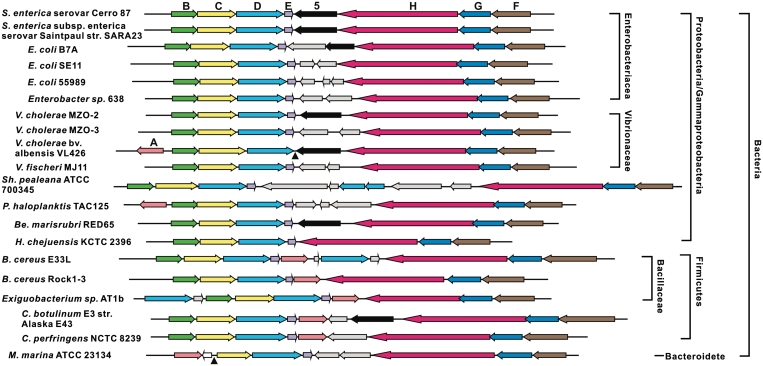
Figure 4.
Organization of 20 bacterial dpt homologs. Colored arrows indicate very similar ORFs. Light gray arrows, diverse ORFs without predicted function that are not homologous to each other. Note that ORF5 (black arrows) is missing from most gene clusters, and dptA homologs (pink arrows) were found in only eight of the clusters. Filled triangles position of presumed deletions in the dptB-F homologous gene clusters. Strains and Genbank accession numbers in the order shown on the figure: S. enterica serovar Cerro 87 (GQ863484); Salmonella enterica subsp. enterica serovar Saintpaul str. SARA23 (NZ_ABAM02000001); E. coli B7A (AAJT02000066); E. coli SE11 (AP009240); E. coli 55989 (CU928145); Enterobacter sp. 638 (CP000653); Vibrio cholerae MZO-2 (AAWF01000002); Vibrio cholerae MZO-3 (AAUU01000003); Vibrio cholerae bv. albensis VL426 (ACHV01000001); Vibrio fischeri MJ11 (CP001133); Shewanella pealeana ATCC 700345 (CP000851); Pseudoalteromonas haloplanktis TAC125 (CR954247); Bermanella marisrubri RED65 (AAQH01000003); Hahella chejuensis KCTC 2396 (CP000155); Bacillus cereus E33L (CP000001); Bacillus cereus Rock1-3 (NZ_ACMG01000043); Exiguobacterium sp. AT1b (CP001615); Clostridium botulinum E3 str. Alaska E43 (CP001078); Clostridium perfringens NCTC 8239 (ABDY01000007); Microscilla marina ATCC 23134 (AAWS00000000).