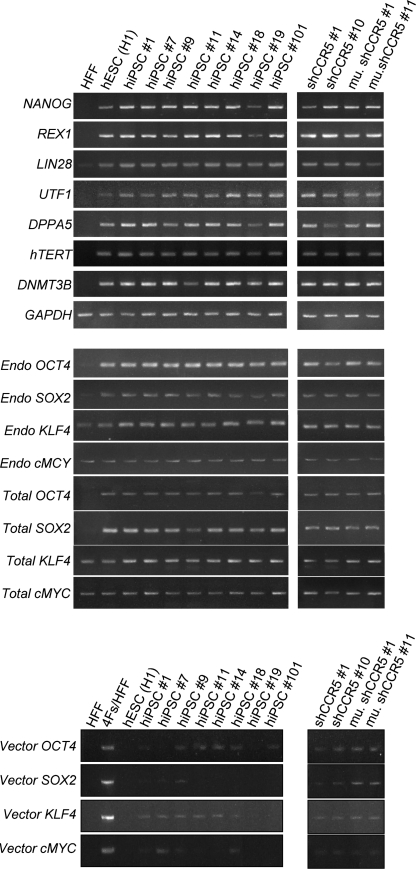
FIG. 5.
Molecular characterization of hiPSCs transduced with or without the lentiviral vector expressing CCR5 shRNA. Total RNA was isolated using QIAGEN's RNeasy Mini kit from untransduced HFFs, HFFs transduced with four reprogramming factors (Oct4, Sox2, Klf4, and cMyc) (4Fs/HFF), hESCs (H1), and hiPSC clones transduced with the lentiviral vector expressing either CCR5 shRNA (sh1005 #1 and #10) or the mutant sh1005 (mu. sh1005 #1 and #11), and hiPSC clones without transduction of any shRNA expressing lentiviral vector (hiPSC #1, #7, #9, #11, #14, #18, #19, and #101). Total RNA (250 ng) was reverse-transcribed using QIAGEN's Omniscript reverse transcription kit and used as a template in subsequent PCR with 5-PRIME's HotMaster Taq DNA polymerase. PCR products were analyzed on a 2% agarose gel. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Primers used for NANOG, REX1, LIN28, UTF1, DPPA5, hTERT, DNMT3B, GAPDH, Endo OCT4, Endo SOX2, Endo KLF4, and Endo cMYC specifically detect the transcripts from the endogenous genes. Primers used for Vector OCT4, Vector SOX2, Vector KLF4, and Vector cMYC specifically detect the transcripts from the vectors used for reprogramming. Primers used for Total OCT4, Total SOX2, Total KLF4, and Total cMYC detect the transcripts from both endogenous and vectors. All primer sequences are listed in Table 1.