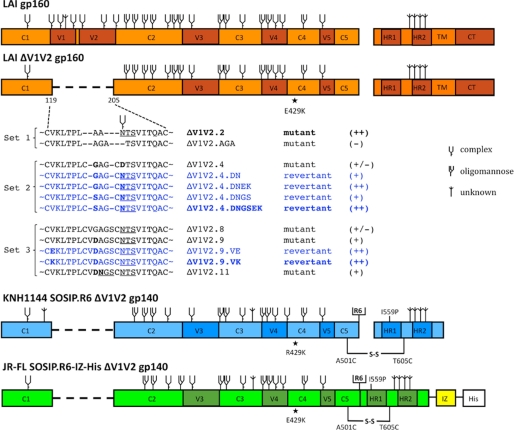
FIGURE 1.
Schematic of ΔV1V2 Env proteins. Constructs based on the sequences from LAI, JR-FL, and KNH1144 are indicated in orange, green, and blue, respectively. Three sets of ΔV1V2 Env proteins were used, including designed mutants (indicated in black) and variants derived from evolution studies (blue). A semi-quantitative assessment of Env function (see Fig. 2) is indicated in braces. Note that the E429K substitution is located far from the site of the V1V2 deletion. The three variants chosen for follow-up analyses are indicated in boldface. The KNH1144 SOSIP.R6 gp140 and JR-FL SOSIP.R6-IZ-His gp140s contain several modifications that have been described elsewhere, including the A501C and T605C substitutions to create the SOS disulfide bond (21), the I559P substitution to promote trimerization (22), and the hexa-arginine (R6) cleavage site to enhance cleavage (47). The SOSIP.R6-IZ-His gp140 also contains the GCN4-based isoleucine zipper (IZ) to promote trimerization (48) and a polyhistidine (His) tag. The N-linked glycan sites on Env are designated as oligomannose or complex, based on experimental determinations using IIIB gp120 (70). It is assumed that the glycans present at analogous sites are processed similarly in Env proteins from other isolates, but note that the carbohydrate composition at some sites has been reported to vary (68–70). Sites that are present on LAI, JR-FL, or KNH1144 gp120, but not on IIIB gp120, or sites that have not been characterized are designated as being of unknown glycan composition.