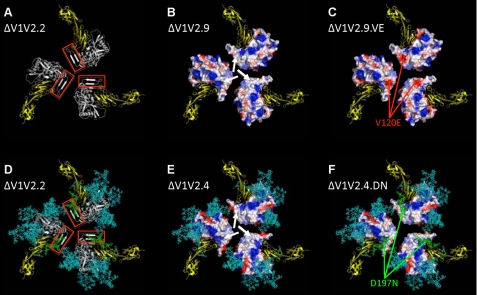
FIGURE 3.
Compensatory mutations affect a hydrophobic surface patch on ΔV1V2 Env trimers. A, ribbon diagram of a ΔV1V2.2 trimer model, depicted from the perspective of the target cell. The gp120 protomers are indicated in white, and the V1V2 stem, part of the bridging sheet, is highlighted by the red box. The two outer domains of CD4 are in yellow. B and C, rendering of the electrostatic surface potentials of the protein-associated, solvent-accessible surfaces of the ΔV1V2.9 and ΔV1V2.9.VE trimers, respectively. Acidic surfaces are in red; basic surfaces are in blue, and neutral surfaces are in white. The white arrows in B indicate a nonpolar surface patch, associated with hydrophobic residues, that is located at the stem of the V1V2 loop structure. The red arrows in C show the negative charge associated with the V120E substitution. D, ribbon diagram of a glycosylated ΔV1V2.2 trimer. The protein is depicted as in A but with the addition of N-glycans, which are represented as sticks in cyan except the one at position 197, which is in green. E and F, rendering of the electrostatic surface potentials of the protein-associated, solvent-accessible surfaces of the ΔV1V2.4 and ΔV1V2.4.DN trimers, respectively. The surface coloration is as in B and C, with the glycans represented as in D. The white arrows in E indicate a neutral surface similar to that shown in B. The N-glycan at residue 197, which partially covers the nonpolar protein-associated surface, is highlighted in green (F).