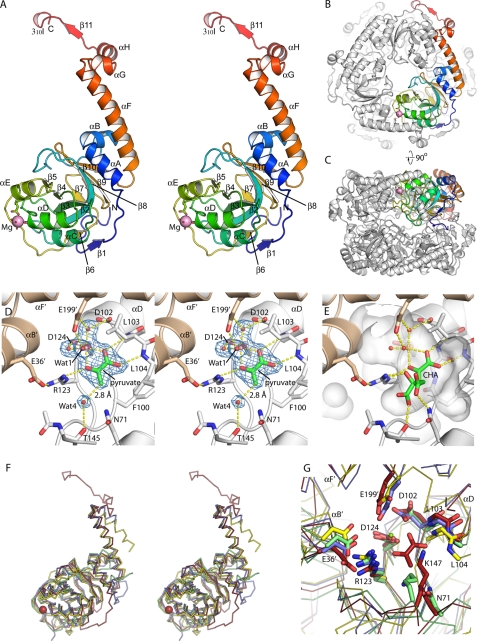
FIGURE 2.
Overall structure of HMG/CHA aldolase. A, organization of the protomer. A single protomer is colored blue to red, N terminus to C terminus. The magnesium ion is shown as a magenta sphere, along with its ligands. Secondary structure elements are labeled. B, view of the hexamer, seen down the 3-fold axis. One protomer is colored as in A, and the others are in white. The C-terminal motif comprising αF, αG, αH, β11, and 310I wrap around the adjacent protomer, forming extensive interactions. C, the hexamer looking down the 2-fold symmetry axis. D, active site of HMG/CHA aldolase. One protomer is colored white, and the other is beige. Electron density for magnesium, pyruvate, and four functionally relevant waters are shown as blue mesh at 1.5 σ. E, model of CHA in the HMG/CHA aldolase binding in the substrate binding pocket (semitransparent white surface). The pyruvate moiety is superposed closely on the experimental pyruvate site. The OH group makes a pair of hydrogen bonds with Wat-1 (the proposed catalytic base) and Arg-123. The carboxylate groups make one direct hydrogen bond each to the protein, with good geometry, as well as to structured water molecules (not shown). F, overlay of HMG/CHA aldolase with its homologs. HMG/CHA aldolase is in red, 3K4I is in blue, 2C5Q is in yellow, and RraA (2PCN) is in green. Note that HMG/CHA aldolase has N- and C-terminal extensions relative to the other proteins, and 2C5Q has a topological inversion relative to the other structures. G, overlay of the catalytic site of HMG/CHA aldolase with its homologs. Structures are colored as in Fig. 2F, residues are labeled by their HMG/CHA aldolase convention. Note that the core catalytic machinery associated with pyruvate binding and activation appears conserved across this whole family.