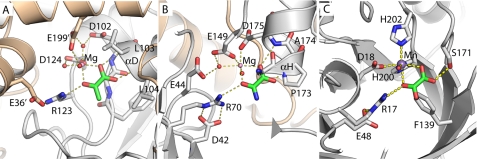
FIGURE 3.
Structural convergence in the active sites of three nonhomologous Class II pyruvate aldolases. A, HMG/CHA aldolase. B, HpaI (Protein Data Bank code 2V5K). C, DmpG (Protein Data Bank code 1NVM). All three structures are presented from the same perspective, taking the metal ion and pyruvate analog as reference. The residues are labeled. For HpaI, the condensing aldehyde will approach from the distal side of the pyruvate, for DmpG and CHA/HMG aldolase from the proximal side. Note that Arg-123–Glu-36′ pair has a direct analog in both HpaI (Arg-70 to Asp-42) and DmpG (Arg-17 to Glu-48), although the nitrogen atom used to hydrogen bond to the keto oxygen is different in each case. HpaI, like HMG/CHA aldolase, uses a magnesium ion coordinated by acidic groups. The similarities with HpaI extend to the αH region, which acts similarly to αD in HMG/CHA aldolase, providing a pair of hydrogen bonds to the oxamate carboxylate group, as well as a metal ligand (here Asp-175). The peptide bond of Gly-172 to Pro-173 acts similarly to the Phe-100 to Gly-101 peptide bond in HMG/CHA aldolase, packing against the side of pyruvate, albeit against the opposite face. The correspondence with DmpG, which uses a manganese ion, is less apparent. The oxalate carboxylate is coordinated by Ser-171 OH instead of using backbone atoms, and Phe-139 packs against the oxalate rather than the backbone immediately N-terminal to the α-helix.