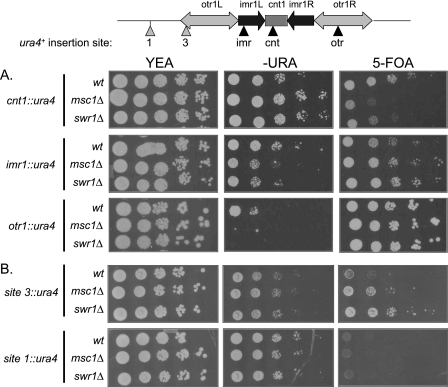
FIGURE 3.
Swr1, like Msc1, affects chromatin structure at the centromere. Strains with ura4+ genes inserted at the locations indicated in the diagram were grown in YEA at 30 °C, and serial dilutions were spotted on YEA plates, minimal media plates lacking uracil, and YEA plates with 5-fluoroorotic acid (5-FOA), which is toxic to cells expressing the ura4+ gene. A, strains with reporters integrated in the centromere of chromosome I. Upper panels, the ura4+ gene is integrated in the central core (cnt1) of the centromere (NW1764, NW1586, and NW2643). Middle panels, the ura4+ gene is integrated at the imr1 region (NW1767, NW1584, and NW2644). Lower panels, the ura4+ gene is integrated in the heterochromatic otr1 region (NW1768, NW1585, and NW2645). B, reporter gene expression at the boundary and outside of the centromere. Upper panels, ura4+ expression at the boundary of otr1 (site 3) was tested in strains NW2648, NW2652, and NW2650. Lower panels, ura4+ expression outside of otr1 (site 1) was tested in strains NW2647, NW2651, and NW2649.