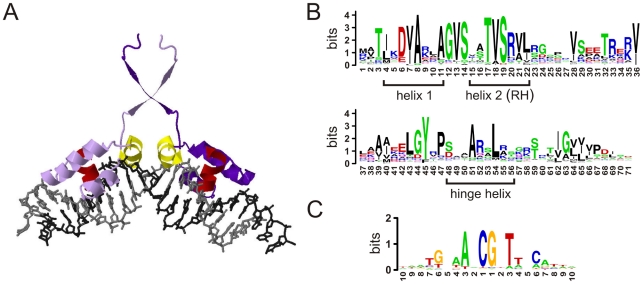
Figure 1. HTH binding mode.
A) X-ray model for a LacI dimer bound to a palindromic BS (plotted with Jmol from the PDB structure 1lbg4). Only the binding domain of each monomer is shown (in light/dark purple, respectively). The hinge-helix and the recognition helix of each monomer are colored in yellow and red, respectively. B) Logo for the alignment of 2639 unredundant HTH-LacI domains. The AA coordinates of any particular domain will be referred by its position in this alignment –they match the numbering of the first 71 AAs of Escherichia coli's GalR and GalS regulators. Helix-1, helix-2 (or recognition helix) and the intermediate residues constitute the HTH motif itself. C) Logo for the alignment of the set of BSs associated to 370 LacI family members (BS sequences from RegTransBase [46]). In BS logos we avoided subscripts for left and right half sites coordinates.