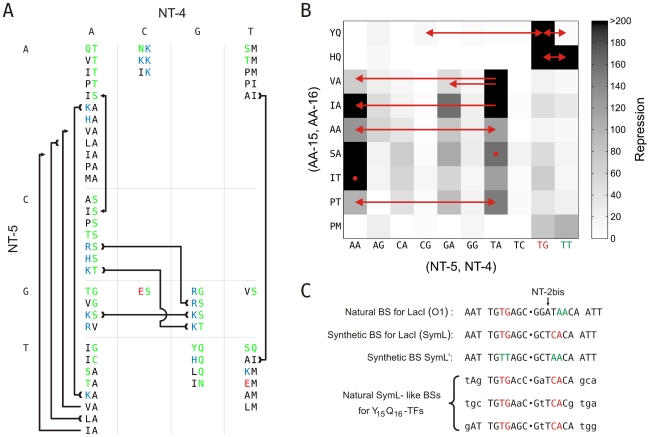
Figure 4. Recognition code and experimental confirmations.
A) Sequence correlations between (AA-15, AA-16) and (NT-5, NT-4) extracted from correlations in Table S1. AAs sequences recognizing a same sequence of NTs were grouped. Here, we only considered significant palindromic NT sequences; for example, (NT-5, NT-4) = TG means 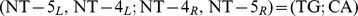 . We included the case for (AA-15, AA-16) = YQ corresponding to the synthetic SymL site in C). Recognition degeneracies are represented as unidirectional arrows (asymmetrical intrinsic), bidirectional divergent arrows (symmetrical intrinsic), and bidirectional convergent arrows (extrinsic). Colors for polar (green), basic (blue), acidic (red) and hydrophobic (black) amino acids. B) Agreement between synthetic and natural data. Recognition of (NT-5, NT-4)-palindromes by different (AA-15, AA-16)-LacI mutants (YQ is the wild type). Data from [19] –from which we only considered those sequences (AA-15, AA-16) with a natural correspondence in Table S1. Rest of BS positions as in SymL. The larger the TF/BS affinity, the stronger the repression of the
. We included the case for (AA-15, AA-16) = YQ corresponding to the synthetic SymL site in C). Recognition degeneracies are represented as unidirectional arrows (asymmetrical intrinsic), bidirectional divergent arrows (symmetrical intrinsic), and bidirectional convergent arrows (extrinsic). Colors for polar (green), basic (blue), acidic (red) and hydrophobic (black) amino acids. B) Agreement between synthetic and natural data. Recognition of (NT-5, NT-4)-palindromes by different (AA-15, AA-16)-LacI mutants (YQ is the wild type). Data from [19] –from which we only considered those sequences (AA-15, AA-16) with a natural correspondence in Table S1. Rest of BS positions as in SymL. The larger the TF/BS affinity, the stronger the repression of the 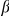 -galactosidase activity. Experimental conditions limited repression to a factor of 200. Arrows indicated again degeneracy classes. Predictions for wild type YQ correspond to asymmetric natural BSs (see text). (NT-5, NT-4)-palindromes involved in the predicted correlations for PM (
-galactosidase activity. Experimental conditions limited repression to a factor of 200. Arrows indicated again degeneracy classes. Predictions for wild type YQ correspond to asymmetric natural BSs (see text). (NT-5, NT-4)-palindromes involved in the predicted correlations for PM (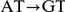 , see Table S1) lack an experimental test. Accordingly, PM do not exhibit a strong affinity for any of the tested palindromes (see Fig. S3), C) Natural and synthetic operators. A dot distinguishes the half sites. Flanking nucleotides separated by a space to help visualization of the highly conserved central region of the BSs. Colors identify different palindromic or mixed combinations in the specificity nucleotides (see Table S2 for more details).
, see Table S1) lack an experimental test. Accordingly, PM do not exhibit a strong affinity for any of the tested palindromes (see Fig. S3), C) Natural and synthetic operators. A dot distinguishes the half sites. Flanking nucleotides separated by a space to help visualization of the highly conserved central region of the BSs. Colors identify different palindromic or mixed combinations in the specificity nucleotides (see Table S2 for more details).