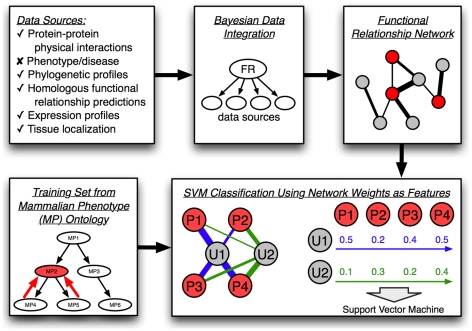
Figure 1. Schematic of our SVM-based functional genomics approach to phenotype prediction.
As shown in the upper half, a functional relationship network was first constructed for the laboratory mouse based on integration of diverse data types, excluding phenotype and disease data to avoid contamination in evaluation. These data were integrated together using Gene Ontology annotations as a gold standard using an established Bayesian pipeline [10]. The resulting network consists of genes as nodes and connections between them representing the probability of two genes participating in the same biological process. This network was used as the basis for an SVM classifier to predict genes associated with phenotypes. As shown in the lower half, annotations to the mammalian phenotype (MP) ontology were used to create gold standards (i.e. training sets) for each SVM. Annotations were propagated along the ontology tree to produce positive training examples (genes associated with the phenotype, labeled P and shown in red). All other genes were considered as unknowns (labeled U and shown in grey). SVMs were provided with input features consisting of network connection weights to positive genes for each phenotype and each SVM was trained to classify unknown examples in this phenotype-specific feature space.