Figure 1.
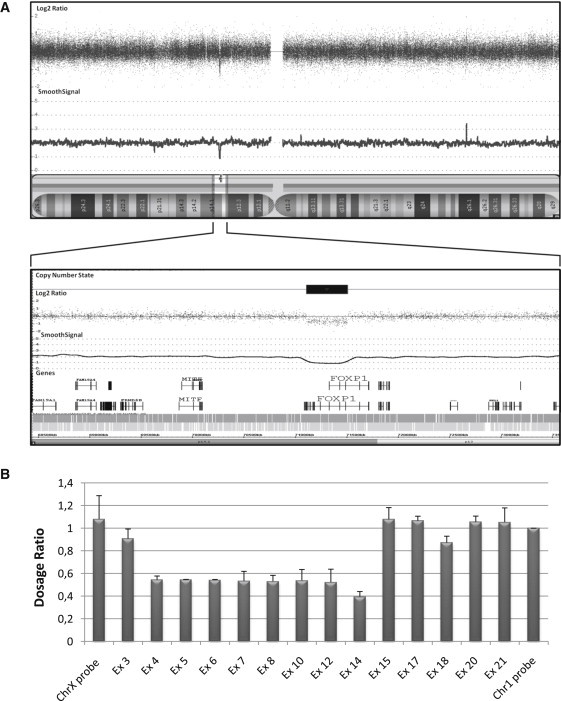
De Novo FOXP1 Deletion at 3p14.1 in Patient A
(A) The Affymetrix Genome-Wide Human SNP 6.0 Array result for patient A is shown above the ideogram of chromosome 3 (Netaffx version 28). Each dot represents a SNP or a copy number marker, with normal copy number having a log2 ratio of ∼0 and deleted regions less than −1. The CNV analysis was performed according to the manufacturer's procedure with the use of the Affymetrix reference library, GenomeWideSNP_6.hapmap270.na30r1.a5, with a regional GC correction. The de novo deletion (chr3: 71109689–71508061; hg18) is boxed, and the only gene affected with this deletion, FOXP1, is shown.
(B) Mapping of FOXP1 deleted exons in patient A. Using MLPA analysis,35 we mapped the deletion breakpoints between exon 4 and exon 14 of FOXP1. MLPA probes targeting 14 exons (Ex) of FOXP1 were custom designed with the ProSeek software.36 These 14 probes were compared to two control probes on chromosome X and chromosome 1. MLPA was performed on 50 ng of genomic DNA. Probe amplification products were run on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The data were analyzed with the GeneMapper software version 4.0 (Applied Biosystems). The dosage ratio (DR) was calculated as follows: DR = (peak height FOXP1 / peak height chromosome 1 probe) in the patient carrying the deletion / (peak height FOXP1 / peak height chromosome 1 probe) in her unaffected mother, who has no copy number variation in FOXP1. The DR has a theoretical value of ≤ 0.7 for a deletion, between 0.7 and 1.3 for a normal situation, and ≥ 1.3 for a duplication. Values represent mean of samples ± standard deviation of samples run in triplicate. Data show that exons 4–14 are deleted in patient A.
