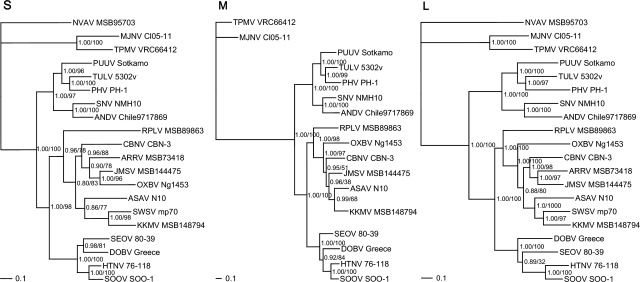
FIG. 2.
Phylogenetic trees generated by the Bayesian method, under the best-fit GTR+I+Γ model of evolution, based on the full-length S and partial M and L genomic segments of Kenkeme virus (KKMV MSB148794) and other selected hantaviruses. The phylogenetic positions of KKMV are shown in relationship to Cao Bang virus (CBNV CBN-3; EF543524, EF543526, EF543525) from the Chinese mole shrew (Anourosorex squamipes), Camp Ripley virus (RPLV MSB89863; FJ790772, EF540774, EF540771) from the northern short-tailed shrew (Blarina brevicauda), Seewis virus (SWSV mp70; EF636024, EF636026) from the Eurasian common shrew (Sorex araneus), Ash River virus (ARRV MSB73418; EF650086, EF619961) from the masked shrew (Sorex cinereus), Jemez Spring virus (JMSV MSB144475; FJ593499, FJ593500, FJ593501) from the dusky shrew (Sorex monticolus), Imjin virus (MJNV Cl05-11; EF641804, EF641798, EF641806) from the Ussuri white-toothed shrew (Crocidura lasiura), Thottapalayam virus (TPMV VRC66412; AY526097, EU001329, EU001330) from the Asian house shrew (Suncus murinus), Asama virus (ASAV N10; EU929072, EU929075, EU929078) from the Japanese shrew mole (Urotrichus talpoides), Oxbow virus (OXBV Ng1453; FJ539166, FJ539167, FJ593497) from the American shrew mole (Neurotrichus gibbsii), and Nova virus (NVAV MSB95703; FJ539168, FJ593498) from the European common mole (Talpa europaea). Also shown are representative rodent-borne hantaviruses, including Hantaan virus (HTNV 76–118; NC_005218, Y00386, NC_005222), Soochong virus (SOOV SOO-1; AY675349, AY675353, DQ056292), Dobrava virus (DOBV Greece; NC_005233, NC_005234, NC_005235), Seoul virus (SEOV 80-39; NC_005236, NC_005237, NC_005238), Tula virus (TULV 5302v; NC_005227, NC_005228, NC_005226), Puumala virus (PUUV Sotkamo; NC_005224, NC_005223, NC_005225), Prospect Hill virus (PHV PH-1; Z49098, X55129, EF646763), Sin Nombre virus (SNV NMH10; NC_005216, NC_005215, NC_005217), and Andes virus (ANDV Chile9717869; NC_003466, NC_003467, NC_003468). GenBank accession numbers are GQ306148, GQ306149, and GQ306150 for the S, M, and L segments of KKMV MSB148794, respectively. SWSV, ARRV, and NVAV are not shown in the M-segment tree because sequences were unavailable for analysis. The numbers at each node are posterior node probabilities (left of slash) based on 30,000 trees, and maximum-likelihood (ML) bootstrap values (right of slash) based on 1000 bootstrap replicates. Bayesian analysis consisted of two replicate Markov Chain Monte Carlo runs of four chains of 2 million generations, each sampled every 100 generations with a burn-in of 5000 (25%). The scale bar indicates nucleotide substitutions per site.